1HJS
 
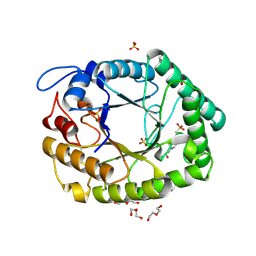 | | Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-1,4-GALACTANASE, ... | | Authors: | Le Nours, J, Ryttersgaard, C, Lo Leggio, L, Ostergaard, P.R, Borchert, T.V, Christensen, L.L.H, Larsen, S. | | Deposit date: | 2003-02-27 | | Release date: | 2003-06-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of Two Fungal Beta-1,4-Galactanases: Searching for the Basis for Temperature and Ph Optimum
Protein Sci., 12, 2003
|
|
1HLW
 
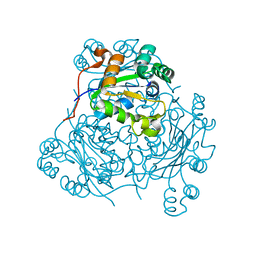 | | STRUCTURE OF THE H122A MUTANT OF THE NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Admiraal, S.J, Meyer, P, Schneider, B, Deville-Bonne, D, Janin, J, Herschlag, D. | | Deposit date: | 2000-12-04 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chemical rescue of phosphoryl transfer in a cavity mutant: a cautionary tale for site-directed mutagenesis.
Biochemistry, 40, 2001
|
|
1ZI6
 
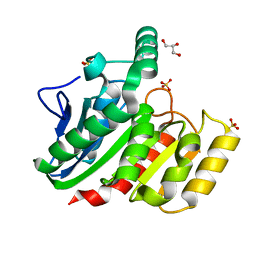 | | Crystal Structure Analysis of the dienelactone hydrolase (C123S) mutant- 1.7 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZIN
 
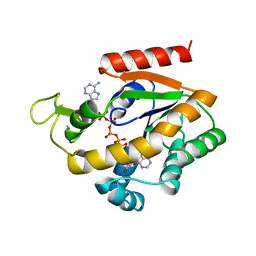 | | ADENYLATE KINASE WITH BOUND AP5A | | Descriptor: | ADENYLATE KINASE, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ZINC ION | | Authors: | Berry, M.B, Phillips Jr, G.N. | | Deposit date: | 1996-06-07 | | Release date: | 1997-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of Bacillus stearothermophilus adenylate kinase with bound Ap5A, Mg2+ Ap5A, and Mn2+ Ap5A reveal an intermediate lid position and six coordinate octahedral geometry for bound Mg2+ and Mn2+.
Proteins, 32, 1998
|
|
1HOE
 
 | |
3GXD
 
 | | Crystal structure of Apo acid-beta-glucosidase pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, PHOSPHATE ION | | Authors: | Lieberman, R.L. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability.
Biochemistry, 48, 2009
|
|
1ZL3
 
 | | Coupling of active site motions and RNA binding | | Descriptor: | 5'-R(*GP*GP*CP*AP*AP*CP*GP*GP*UP*(FLO) UP*CP*GP*AP*UP*CP*CP*CP*GP*UP*UP*GP*C)-3', SULFATE ION, tRNA pseudouridine synthase B | | Authors: | Hoang, C, Hamilton, C.S, Mueller, E.G, Ferre-D'Amare, A.R. | | Deposit date: | 2005-05-05 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Precursor complex structure of pseudouridine synthase TruB suggests coupling of active site perturbations to an RNA-sequestering peripheral protein domain
Protein Sci., 14, 2005
|
|
1HWH
 
 | |
1Z9J
 
 | | Photosynthetic Reaction Center from Rhodobacter sphaeroides | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Thielges, M, Uyeda, G, Camara-Artigas, A, Kalman, L, Williams, J.C, Allen, J.P. | | Deposit date: | 2005-04-02 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Design of a Redox-Linked Active Metal Site: Manganese Bound to Bacterial Reaction Centers at a Site Resembling That of Photosystem II
Biochemistry, 44, 2005
|
|
1HIM
 
 | |
1HJB
 
 | |
1ZNS
 
 | | Crystal structure of N-ColE7/12-bp DNA/Zn complex | | Descriptor: | 5'-D(*CP*GP*GP*GP*AP*TP*AP*TP*CP*CP*CP*G)-3', Colicin E7, ZINC ION | | Authors: | Doudeva, L.G, Huang, H, Hsia, K.C, Shi, Z, Li, C.L, Shen, Y, Yuan, H.S. | | Deposit date: | 2005-05-12 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structural analysis and metal-dependent stability and activity studies of the ColE7 endonuclease domain in complex with DNA/Zn2+ or inhibitor/Ni2+
Protein Sci., 15, 2006
|
|
1ZP4
 
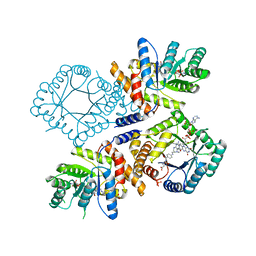 | | Glu28Gln mutant of E. coli Methylenetetrahydrofolate Reductase (oxidized) complex with Methyltetrahydrofolate | | Descriptor: | 5,10-methylenetetrahydrofolate reductase, 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Pejchal, R, Sargeant, R, Ludwig, M.L. | | Deposit date: | 2005-05-16 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of NADH and CH(3)-H(4)Folate Complexes of Escherichia coli Methylenetetrahydrofolate Reductase Reveal a Spartan Strategy for a Ping-Pong Reaction
Biochemistry, 44, 2005
|
|
1HUY
 
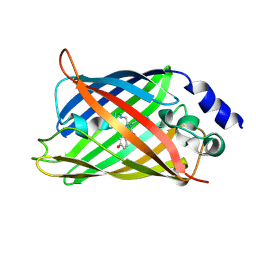 | | CRYSTAL STRUCTURE OF CITRINE, AN IMPROVED YELLOW VARIANT OF GREEN FLUORESCENT PROTEIN | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Griesbeck, O, Baird, G.S, Campbell, R.E, Zacharias, D.A, Tsien, R.Y. | | Deposit date: | 2001-01-04 | | Release date: | 2001-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications
J.Biol.Chem., 276, 2001
|
|
1HLU
 
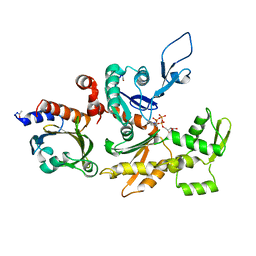 | | STRUCTURE OF BOVINE BETA-ACTIN-PROFILIN COMPLEX WITH ACTIN BOUND ATP PHOSPHATES SOLVENT ACCESSIBLE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, BETA-ACTIN, CALCIUM ION, ... | | Authors: | Chik, J.K, Lindberg, U, Schutt, C.E. | | Deposit date: | 1997-05-30 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The structure of an open state of beta-actin at 2.65 A resolution.
J.Mol.Biol., 263, 1996
|
|
1HNY
 
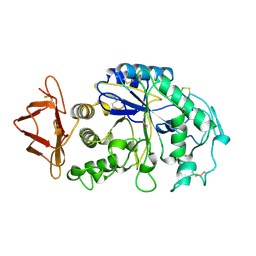 | |
1HRS
 
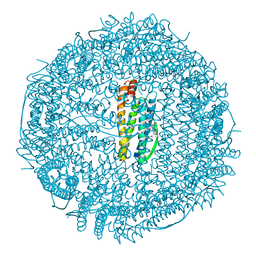 | | A CRYSTALLOGRAPHIC STUDY OF HAEM BINDING TO FERRITIN | | Descriptor: | APOFERRITIN CO-CRYSTALLIZED WITH SN-PROTOPORPHYRIN IX IN CADMIUM SULFATE, CADMIUM ION, PROTOPORPHYRIN IX | | Authors: | Precigoux, G, Yariv, J, Gallois, B, Dautant, A, Courseille, C, Langlois D'Estaintot, B. | | Deposit date: | 1993-11-05 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A crystallographic study of haem binding to ferritin.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1HSI
 
 | |
1ZG7
 
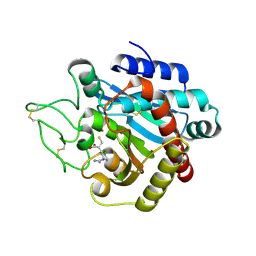 | | Crystal Structure of 2-(5-{[amino(imino)methyl]amino}-2-chlorophenyl)-3-sulfanylpropanoic acid Bound to Activated Porcine Pancreatic Carboxypeptidase B | | Descriptor: | 2-(5-{[AMINO(IMINO)METHYL]AMINO}-2-CHLOROPHENYL)-3-SULFANYLPROPANOIC ACID, ZINC ION, procarboxypeptidase B | | Authors: | Adler, M, Bryant, J, Buckman, B, Islam, I, Larsen, B, Finster, S, Kent, L, May, K, Mohan, R, Yuan, S, Whitlow, M. | | Deposit date: | 2005-04-20 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of potent thiol-based inhibitors bound to carboxypeptidase b.
Biochemistry, 44, 2005
|
|
1HSG
 
 | |
1HMK
 
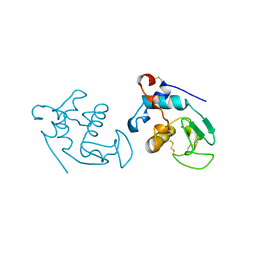 | | RECOMBINANT GOAT ALPHA-LACTALBUMIN | | Descriptor: | CALCIUM ION, PROTEIN (ALPHA-LACTALBUMIN) | | Authors: | Horii, K, Matsushima, M, Tsumoto, K, Kumagai, I. | | Deposit date: | 1998-11-26 | | Release date: | 1999-11-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effect of the extra n-terminal methionine residue on the stability and folding of recombinant alpha-lactalbumin expressed in Escherichia coli.
J.Mol.Biol., 285, 1999
|
|
1HOP
 
 | | STRUCTURE OF GUANINE NUCLEOTIDE (GPPCP) COMPLEX OF ADENYLOSUCCINATE SYNTHETASE FROM ESCHERICHIA COLI AT PH 6.5 AND 25 DEGREES CELSIUS | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Poland, B.W, Hou, Z, Bruns, C, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1996-04-26 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined crystal structures of guanine nucleotide complexes of adenylosuccinate synthetase from Escherichia coli.
J.Biol.Chem., 271, 1996
|
|
1HOC
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF H-2DB AT 2.4 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ANTIGEN-DETERMINANT SELECTION | | Descriptor: | 9-RESIDUE PEPTIDE, BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (H2-DB) (ALPHA CHAIN) | | Authors: | Young, A.C.M, Zhang, W, Sacchettini, J.C. | | Deposit date: | 1994-01-02 | | Release date: | 1994-04-30 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure of H-2Db at 2.4 A resolution: implications for antigen-determinant selection
Cell(Cambridge,Mass.), 76, 1994
|
|
1ZHP
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with Benzamidine (S434A-T475A-K505 Mutant) | | Descriptor: | BENZAMIDINE, GLUTATHIONE, coagulation factor XI | | Authors: | Jin, L, Pandey, P, Babine, R.E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2005-04-26 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mutation of surface residues to promote crystallization of activated factor XI as a complex with benzamidine: an essential step for the iterative structure-based design of factor XI inhibitors.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZHZ
 
 | | Structure of yeast oxysterol binding protein Osh4 in complex with ergosterol | | Descriptor: | ERGOSTEROL, KES1 protein, LEAD (II) ION | | Authors: | Im, Y.J, Raychaudhuri, S, Prinz, W.A, Hurley, J.H. | | Deposit date: | 2005-04-26 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural mechanism for sterol sensing and transport by OSBP-related proteins
Nature, 437, 2005
|
|
