7VAO
 
 | | V1EG of V/A-ATPase from Thermus thermophilus, high ATP, state2-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kishikawa, J, Nakanishi, A, Nakano, A, Saeki, S, Furuta, A, Kato, T, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2021-08-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases.
Nat Commun, 13, 2022
|
|
7VAR
 
 | | V1EG domain of V/A-ATPase from Thermus thermophilus at low ATP concentration, state1-1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kishikawa, J, Nakanishi, A, Nakano, A, Saeki, S, Furuta, A, Kato, T, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2021-08-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases.
Nat Commun, 13, 2022
|
|
7VAS
 
 | | V1EG domain of V/A-ATPase from Thermus thermophilus at low ATP concentration, state1-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kishikawa, J, Nakanishi, A, Nakano, A, Saeki, S, Furuta, A, Kato, T, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2021-08-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases.
Nat Commun, 13, 2022
|
|
7VAV
 
 | | V1EG of V/A-ATPase from Thermus thermophilus at low ATP concentration, state3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kishikawa, J, Nakanishi, A, Nakano, A, Saeki, S, Furuta, A, Kato, T, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2021-08-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases.
Nat Commun, 13, 2022
|
|
7VAU
 
 | | V1EG of V/A-ATPase from Thermus thermophilus at low ATP concentration, state2-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kishikawa, J, Nakanishi, A, Nakano, A, Saeki, S, Furuta, A, Kato, T, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2021-08-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases.
Nat Commun, 13, 2022
|
|
7VAY
 
 | | V1EG domain of V/A-ATPase from Thermus thermophilus at saturated ATP-gamma-S condition, state2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Kishikawa, J, Nakanishi, A, Nakano, A, Saeki, S, Furuta, A, Kato, T, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2021-08-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases.
Nat Commun, 13, 2022
|
|
6QUM
 
 | |
6R0W
 
 | |
6R0Z
 
 | |
6R0Y
 
 | |
6R10
 
 | |
8YXZ
 
 | | Vo domain of V/A-ATPase from Thermus thermophilus state1 | | Descriptor: | V-type ATP synthase subunit C, V-type ATP synthase subunit I, V-type ATP synthase, ... | | Authors: | Kishikawa, J, Nishida, Y, Nakano, A, Yokoyama, K. | | Deposit date: | 2024-04-03 | | Release date: | 2024-12-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Rotary mechanism of the prokaryotic V o motor driven by proton motive force.
Nat Commun, 15, 2024
|
|
8YY0
 
 | | Vo domain of V/A-ATPase from Thermus thermophilus state2 | | Descriptor: | V-type ATP synthase subunit C, V-type ATP synthase subunit I, V-type ATP synthase, ... | | Authors: | Kishikawa, J, Nishida, Y, Nakano, A, Yokoyama, K. | | Deposit date: | 2024-04-03 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Rotary mechanism of the prokaryotic V o motor driven by proton motive force.
Nat Commun, 15, 2024
|
|
8YY1
 
 | | Vo domain of V/A-ATPase from Thermus thermophilus state3 | | Descriptor: | V-type ATP synthase subunit C, V-type ATP synthase subunit I, V-type ATP synthase, ... | | Authors: | Nishida, Y, Kishikawa, J, Nakano, A, Yokoyama, K. | | Deposit date: | 2024-04-03 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Rotary mechanism of the prokaryotic V o motor driven by proton motive force.
Nat Commun, 15, 2024
|
|
3SDZ
 
 | | Structural characterization of the subunit A mutant F427W of the A-ATP synthase from Pyrococcus horikoshii | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Tadwal, V.S, Manimekalai, M.S.S, Balakrishna, A.M, Gruber, G. | | Deposit date: | 2011-06-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Engineered tryptophan in the adenine-binding pocket of catalytic subunit A of A-ATP synthase demonstrates the importance of aromatic residues in adenine binding, forming a tool for steady-state and time-resolved fluorescence spectroscopy.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2KWY
 
 | | Structure of G61-101 | | Descriptor: | V-type proton ATPase subunit G | | Authors: | Rishikesan, S, Gruber, G. | | Deposit date: | 2010-04-22 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of G61-101
To be Published
|
|
6KOJ
 
 | | Crystal structure of SNX11-PXe domain in complex with PI(3,5)P2 | | Descriptor: | Sorting nexin-11, [(2~{R})-2-butanoyloxy-3-[oxidanyl-[(2~{R},3~{R},5~{S},6~{R})-2,4,6-tris(oxidanyl)-3,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] butanoate | | Authors: | Xu, T, Liu, J. | | Deposit date: | 2019-08-11 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Molecular Basis for PI(3,5)P2Recognition by SNX11, a Protein Involved in Lysosomal Degradation and Endosome Homeostasis Regulation.
J.Mol.Biol., 432, 2020
|
|
6KOK
 
 | | Crystal Structure of SNX11/SNX10-PXe Chimera | | Descriptor: | CHLORIDE ION, SODIUM ION, Sorting nexin-11,Uncharacterized protein SNX10 | | Authors: | Xu, T, Liu, J. | | Deposit date: | 2019-08-11 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Basis for PI(3,5)P2Recognition by SNX11, a Protein Involved in Lysosomal Degradation and Endosome Homeostasis Regulation.
J.Mol.Biol., 432, 2020
|
|
6KOI
 
 | | Crystal structure of SNX11-PXe domain in dimer form. | | Descriptor: | Sorting nexin-11 | | Authors: | Xu, T, Xu, J, Liu, J. | | Deposit date: | 2019-08-11 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular Basis for PI(3,5)P2Recognition by SNX11, a Protein Involved in Lysosomal Degradation and Endosome Homeostasis Regulation.
J.Mol.Biol., 432, 2020
|
|
6F36
 
 | | Polytomella Fo model | | Descriptor: | Mitochondrial ATP synthase subunit 6, Mitochondrial ATP synthase subunit ASA6, Mitochondrial ATP synthase subunit c | | Authors: | Yildiz, O, Kuehlbrandt, W, Klusch, N, Murphy, B.J, Mills, D.J. | | Deposit date: | 2017-11-28 | | Release date: | 2017-12-20 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of proton translocation and force generation in mitochondrial ATP synthase.
Elife, 6, 2017
|
|
8J0T
 
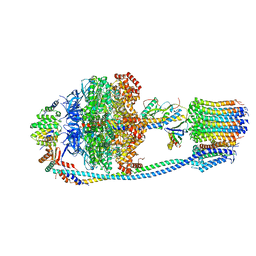 | | Cryo-EM structure of Mycobacterium tuberculosis ATP synthase in the apo-form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Zhang, Y, Lai, Y, Liu, F, Rao, Z, Gong, H. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Inhibition of M. tuberculosis and human ATP synthase by BDQ and TBAJ-587.
Nature, 631, 2024
|
|
4BEM
 
 | | Crystal structure of the F-type ATP synthase c-ring from Acetobacterium woodii. | | Descriptor: | ACETATE ION, F1FO ATPASE C1 SUBUNIT, F1FO ATPASE C2 SUBUNIT, ... | | Authors: | Matthies, D, Meier, T, Yildiz, O. | | Deposit date: | 2013-03-11 | | Release date: | 2014-03-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-Resolution Structure and Mechanism of an F/V-Hybrid Rotor Ring in a Na+-Coupled ATP Synthase
Nat.Commun., 5, 2014
|
|
8SXR
 
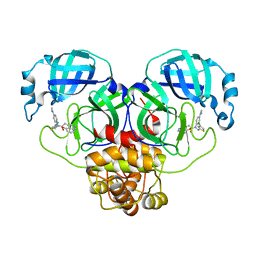 | | Crystal structure of SARS-CoV-2 Mpro with C5a | | Descriptor: | 3C-like proteinase nsp5, N-[(4-chlorothiophen-2-yl)methyl]-N-[4-(dimethylamino)phenyl]-2-(5-hydroxyisoquinolin-4-yl)acetamide | | Authors: | Worrall, L.J, Kenward, C, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2023-05-23 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
2WIE
 
 | | High-resolution structure of the rotor ring from a proton dependent ATP synthase | | Descriptor: | ATP SYNTHASE C CHAIN, CYMAL-4 | | Authors: | Pogoryelov, D, Yildiz, O, Faraldo-Gomez, J.D, Meier, T. | | Deposit date: | 2009-05-11 | | Release date: | 2009-09-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | High-Resolution Structure of the Rotor Ring of a Proton-Dependent ATP Synthase.
Nat.Struct.Mol.Biol., 16, 2009
|
|
4O1S
 
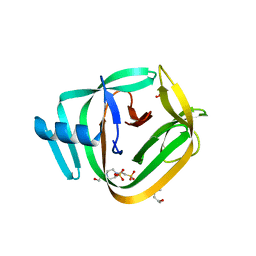 | | Crystal structure of TvoVMA intein | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, ... | | Authors: | Aranko, A.S, Oeemig, J.S, Zhou, D, Kajander, T, Wlodawer, A, Iwai, H. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7008 Å) | | Cite: | Structure-based engineering and comparison of novel split inteins for protein ligation.
Mol Biosyst, 10, 2014
|
|
