1KBP
 
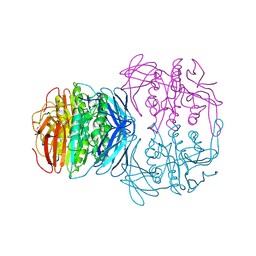 | | KIDNEY BEAN PURPLE ACID PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, PURPLE ACID PHOSPHATASE, ... | | Authors: | Klabunde, T, Strater, N, Krebs, B. | | Deposit date: | 1995-02-20 | | Release date: | 1996-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanism of Fe(III)-Zn(II) purple acid phosphatase based on crystal structures.
J.Mol.Biol., 259, 1996
|
|
1FBD
 
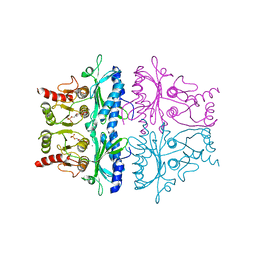 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, FRUCTOSE 1,6-BISPHOSPHATASE, MANGANESE (II) ION | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
1FBU
 
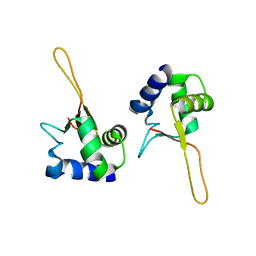 | | HEAT SHOCK TRANSCRIPTION FACTOR DNA BINDING DOMAIN | | Descriptor: | HEAT SHOCK FACTOR PROTEIN | | Authors: | Hardy, J.A, Nelson, H.C.M. | | Deposit date: | 2000-07-16 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Proline in alpha-helical kink is required for folding kinetics but not for kinked structure, function, or stability of heat shock transcription factor.
Protein Sci., 9, 2000
|
|
1FRQ
 
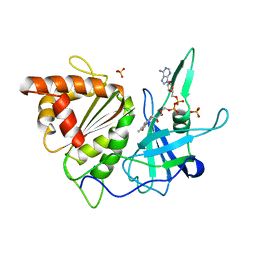 | | FERREDOXIN:NADP+ OXIDOREDUCTASE (FERREDOXIN REDUCTASE) MUTANT E312A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, PROTEIN (FERREDOXIN:NADP+ OXIDOREDUCTASE), ... | | Authors: | Aliverti, A, Deng, Z, Ravasi, D, Piubelli, L, Karplus, P.A, Zanetti, G. | | Deposit date: | 1998-10-10 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the function of the invariant glutamyl residue 312 in spinach ferredoxin-NADP+ reductase.
J.Biol.Chem., 273, 1998
|
|
1FGV
 
 | |
1FH8
 
 | | CRYSTAL STRUCTURE OF THE XYLANASE CEX WITH XYLOBIOSE-DERIVED ISOFAGOMINE INHIBITOR | | Descriptor: | BETA-1,4-XYLANASE, PIPERIDINE-3,4-DIOL, beta-D-xylopyranose | | Authors: | Notenboom, V, Williams, S.J, Hoos, R, Withers, S.G, Rose, D.R. | | Deposit date: | 2000-07-31 | | Release date: | 2000-08-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Detailed structural analysis of glycosidase/inhibitor interactions: complexes of Cex from Cellulomonas fimi with xylobiose-derived aza-sugars.
Biochemistry, 39, 2000
|
|
1FAJ
 
 | | INORGANIC PYROPHOSPHATASE | | Descriptor: | SOLUBLE INORGANIC PYROPHOSPHATASE | | Authors: | Kankare, J.A, Salminen, T, Goldman, A. | | Deposit date: | 1996-01-10 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Escherichia coli inorganic pyrophosphatase at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1FIQ
 
 | | CRYSTAL STRUCTURE OF XANTHINE OXIDASE FROM BOVINE MILK | | Descriptor: | 2-HYDROXYBENZOIC ACID, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Enroth, C, Eger, B.T, Okamoto, K, Nishino, T, Nishino, T, Pai, E.F. | | Deposit date: | 2000-08-04 | | Release date: | 2000-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FEC
 
 | |
1KB0
 
 | |
1FL0
 
 | | CRYSTAL STRUCTURE OF THE EMAP2/RNA-BINDING DOMAIN OF THE P43 PROTEIN FROM HUMAN AMINOACYL-TRNA SYNTHETASE COMPLEX | | Descriptor: | ENDOTHELIAL-MONOCYTE ACTIVATING POLYPEPTIDE II | | Authors: | Renault, L, Kerjan, P, Pasqualato, S, Menetrey, J, Robinson, J.-C, Kawaguchi, S, Vassylyev, D.G, Yokoyama, S, Mirande, M, Cherfils, J. | | Deposit date: | 2000-08-11 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the EMAPII domain of human aminoacyl-tRNA synthetase complex reveals evolutionary dimer mimicry.
EMBO J., 20, 2001
|
|
1FLP
 
 | | STRUCTURE OF THE SULFIDE-REACTIVE HEMOGLOBIN FROM THE CLAM LUCINA PECTINATA: CRYSTALLOGRAPHIC ANALYSIS AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | HEMOGLOBIN I (AQUO MET), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Rizzi, M, Wittenberg, J.B, Ascenzi, P, Fasano, M, Coda, A, Bolognesi, M. | | Deposit date: | 1994-05-16 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the sulfide-reactive hemoglobin from the clam Lucina pectinata. Crystallographic analysis at 1.5 A resolution.
J.Mol.Biol., 244, 1994
|
|
1FLV
 
 | |
1KMB
 
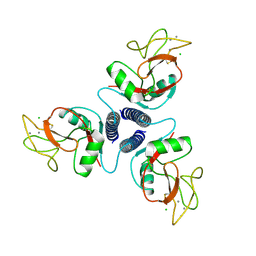 | | SELECTIN-LIKE MUTANT OF MANNOSE-BINDING PROTEIN A | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-A | | Authors: | Ng, K.K.-S, Weis, W.I. | | Deposit date: | 1996-11-07 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a selectin-like mutant of mannose-binding protein complexed with sialylated and sulfated Lewis(x) oligosaccharides.
Biochemistry, 36, 1997
|
|
1KCA
 
 | |
1ICJ
 
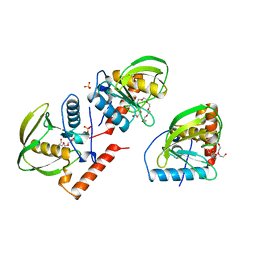 | | PDF PROTEIN IS CRYSTALLIZED AS NI2+ CONTAINING FORM, COCRYSTALLIZED WITH INHIBITOR POLYETHYLENE GLYCOL (PEG) | | Descriptor: | NICKEL (II) ION, NONAETHYLENE GLYCOL, PEPTIDE DEFORMYLASE, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-03-12 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of peptide deformylase and identification of the substrate binding site.
J.Biol.Chem., 273, 1998
|
|
1FO8
 
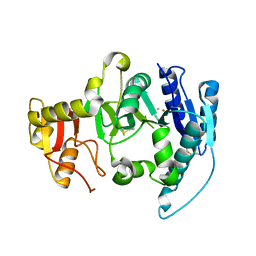 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINYLTRANSFERASE I | | Descriptor: | ALPHA-1,3-MANNOSYL-GLYCOPROTEIN BETA-1,2-N-ACETYLGLUCOSAMINYLTRANSFERASE, METHYL MERCURY ION | | Authors: | Unligil, U.M, Zhou, S, Yuwaraj, S, Sarkar, M, Schachter, H, Rini, J.M. | | Deposit date: | 2000-08-26 | | Release date: | 2001-04-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of rabbit N-acetylglucosaminyltransferase I: catalytic mechanism and a new protein superfamily.
EMBO J., 19, 2000
|
|
1FOJ
 
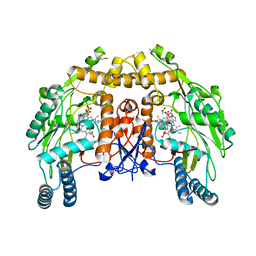 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 7-NITROINDAZOLE-2-CARBOXAMIDINE (H4B PRESENT) | | Descriptor: | 7-NITROINDAZOLE, 7-NITROINDAZOLE-2-CARBOXAMIDINE, ACETATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Masters, B.S, Poulos, T.L. | | Deposit date: | 2000-08-28 | | Release date: | 2001-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of nitric oxide synthase bound to nitro indazole reveals a novel inactivation mechanism
Biochemistry, 40, 2001
|
|
1FQI
 
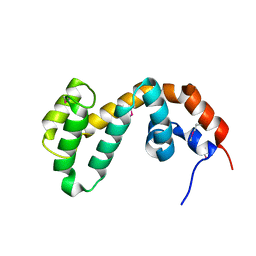 | | RGS9 RGS DOMAIN | | Descriptor: | REGULATOR OF G-PROTEIN SIGNALING 9 | | Authors: | Slep, K.C, Kercher, M.A, He, W, Cowan, C.W, Wensel, T.G, Sigler, P.B. | | Deposit date: | 2000-09-05 | | Release date: | 2001-02-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural determinants for regulation of phosphodiesterase by a G protein at 2.0 A.
Nature, 409, 2001
|
|
1FGI
 
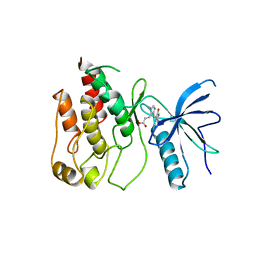 | |
1FOO
 
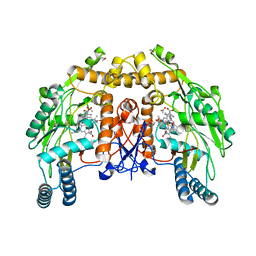 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH L-ARG AND NO(H4B-FREE) | | Descriptor: | ACETATE ION, ARGININE, CACODYLATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 2000-08-28 | | Release date: | 2001-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies on endothelial nitric oxide synthase complexed with nitric oxide and mechanism-based inhibitors.
Biochemistry, 40, 2001
|
|
1F8V
 
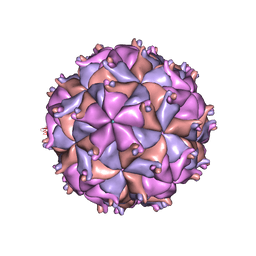 | | THE STRUCTURE OF PARIACOTO VIRUS REVEALS A DODECAHEDRAL CAGE OF DUPLEX RNA | | Descriptor: | CALCIUM ION, MATURE CAPSID PROTEIN BETA, MATURE CAPSID PROTEIN GAMMA, ... | | Authors: | Tang, L, Johnson, K.N, Ball, L.A, Lin, T, Yeager, M, Johnson, J.E. | | Deposit date: | 2000-07-05 | | Release date: | 2000-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of pariacoto virus reveals a dodecahedral cage of duplex RNA.
Nat.Struct.Biol., 8, 2001
|
|
1FH7
 
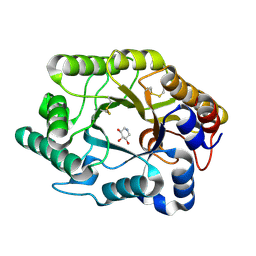 | | CRYSTAL STRUCTURE OF THE XYLANASE CEX WITH XYLOBIOSE-DERIVED INHIBITOR DEOXYNOJIRIMYCIN | | Descriptor: | BETA-1,4-XYLANASE, PIPERIDINE-3,4,5-TRIOL, beta-D-xylopyranose | | Authors: | Notenboom, V, Williams, S.J, Hoos, R, Withers, S.G, Rose, D.R. | | Deposit date: | 2000-07-31 | | Release date: | 2000-08-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Detailed structural analysis of glycosidase/inhibitor interactions: complexes of Cex from Cellulomonas fimi with xylobiose-derived aza-sugars.
Biochemistry, 39, 2000
|
|
1I2P
 
 | |
1FQW
 
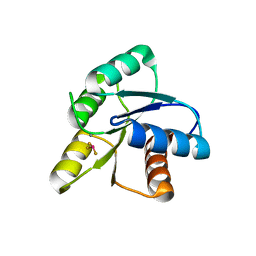 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, MANGANESE (II) ION | | Authors: | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Berry, E.A, Wemmer, D.E. | | Deposit date: | 2000-09-07 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of activated CheY. Comparison with other activated receiver domains.
J.Biol.Chem., 276, 2001
|
|
