6GMS
 
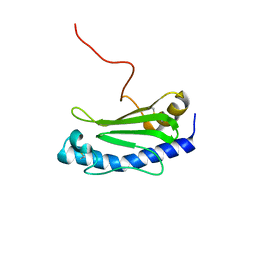 | | Solution NMR structure of the major type IV pilin PpdD from enterohemorrhagic Escherichia coli (EHEC) | | Descriptor: | Prepilin peptidase-dependent protein D | | Authors: | Amorim, G.C, Bardiaux, B, Luna-Rico, A, Zeng, W, Guilvout, I, Egelman, E, Nilges, M, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2018-05-28 | | Release date: | 2019-05-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and Assembly of the Enterohemorrhagic Escherichia coli Type 4 Pilus.
Structure, 27, 2019
|
|
2Z33
 
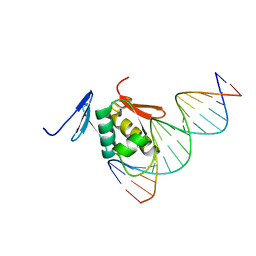 | | Solution structure of the DNA complex of PhoB DNA-binding/transactivation Domain | | Descriptor: | 5'-D(*AP*CP*AP*GP*AP*TP*TP*TP*AP*TP*GP*AP*CP*AP*GP*T)-3', 5'-D(*AP*CP*TP*GP*TP*CP*AP*TP*AP*AP*AP*TP*CP*TP*GP*T)-3', Phosphate regulon transcriptional regulatory protein phoB | | Authors: | Yamane, T, Okamura, H, Ikeguchi, M, Nishimura, Y, Kidera, A. | | Deposit date: | 2007-05-31 | | Release date: | 2008-04-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Water-mediated interactions between DNA and PhoB DNA-binding/transactivation domain: NMR-restrained molecular dynamics in explicit water environment.
Proteins, 71, 2008
|
|
7P51
 
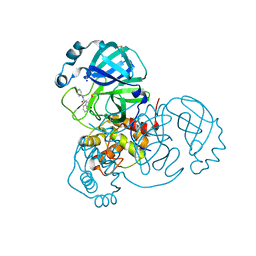 | | CRYSTAL STRUCTURE OF THE SARS-COV-2 MAIN PROTEASE COMPLEXED WITH FRAGMENT F01 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(5-chloropyridin-2-yl)-3-oxo-2,3-dihydro-1H-indene-1-carboxamide, ... | | Authors: | Hanoulle, X, Moschidi, D. | | Deposit date: | 2021-07-13 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.474 Å) | | Cite: | NMR Spectroscopy of the Main Protease of SARS-CoV-2 and Fragment-Based Screening Identify Three Protein Hotspots and an Antiviral Fragment.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5O9B
 
 | |
6QTF
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring B, major conformer | | Descriptor: | DCY-LEU-GLY-ALA-THR | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-02-25 | | Release date: | 2019-09-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6QM1
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Nisin Ring B (Lan8,11) analogue | | Descriptor: | DAL-PRO-GLY-CYS-LYS | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-02-01 | | Release date: | 2019-09-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6S0N
 
 | | A9 peptide derived from Herceptin fab binding region | | Descriptor: | GLN-ASP-VAL-ASN-THR-ALA-VAL-ALA-TRP | | Authors: | De Luca, S, Verdoliva, V, Saviano, M, Fattorusso, R, Diana, D. | | Deposit date: | 2019-06-17 | | Release date: | 2019-11-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | SPR and NMR characterization of the molecular interaction between A9 peptide and a model system of HER2 receptor: A fragment approach for selecting peptide structures specific for their target.
J.Pept.Sci., 26, 2020
|
|
7WCG
 
 | |
7VUT
 
 | | Crystal structure of AlleyCat10 | | Descriptor: | AlleyCat10, CALCIUM ION | | Authors: | Tame, J.R.H, Korendovych, I.V, Margheritis, E, Takahashi, K. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | NMR-guided directed evolution.
Nature, 610, 2022
|
|
7VUS
 
 | | Crystal structure of AlleyCat9 with 5-nitro-benzotriazole | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-nitro-1H-benzotriazole, AlleyCat, ... | | Authors: | Tame, J.R.H, Korendovych, I.V, Margheritis, E, Takahashi, K. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | NMR-guided directed evolution.
Nature, 610, 2022
|
|
7VUR
 
 | | Crystal structure of AlleyCat9 with calcium but no inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, AlleyCat, CALCIUM ION | | Authors: | Margheritis, E, Takahashi, K, Korendovych, I.V, Tame, J.R.H. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | NMR-guided directed evolution.
Nature, 610, 2022
|
|
7VUC
 
 | |
7VUU
 
 | | Crystal structure of AlleyCat10 with inhibitor | | Descriptor: | 5-nitro-1H-benzotriazole, AlleyCat, CALCIUM ION | | Authors: | Tame, J.R.H, Korendovych, I.V, Margheritis, E, Takahashi, K. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | NMR-guided directed evolution.
Nature, 610, 2022
|
|
7TB9
 
 | | Structural characterization of the biological synthetic peptide pCEMP1 | | Descriptor: | CEMP1-p1 | | Authors: | Lopez Giraldo, A, del Rio Portilla, F, Nidome Campos, M, Romo Arevalo, E, Arzate, H. | | Deposit date: | 2021-12-21 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of cementum protein 1 derived peptide (CEMP1-p1) and its role in the mineralization process.
J.Pept.Sci., 29, 2023
|
|
6ZFV
 
 | |
2YTX
 
 | | Solution structure of the second cold-shock domain of the human KIAA0885 protein (UNR protein) | | Descriptor: | Cold shock domain-containing protein E1 | | Authors: | Goroncy, A.K, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
2YTV
 
 | | Solution structure of the fifth cold-shock domain of the human KIAA0885 protein (unr protein) | | Descriptor: | Cold shock domain-containing protein E1 | | Authors: | Goroncy, A.K, Tochio, N, Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
5LCH
 
 | | VIM-2 metallo-beta-lactamase in complex with (S)-1-allyl-2-(3-methoxyphenyl)-3-oxoisoindoline-4-carboxylic acid (compound 42) | | Descriptor: | (1~{S})-2-(3-methoxyphenyl)-3-oxidanylidene-1-prop-2-enyl-1~{H}-isoindole-4-carboxylic acid, Metallo-beta-lactamase VIM-2, ZINC ION | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2016-06-21 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | NMR-filtered virtual screening leads to non-metal chelating metallo-beta-lactamase inhibitors.
Chem Sci, 8, 2017
|
|
5LCA
 
 | | VIM-2 metallo-beta-lactamase in complex with 3-oxo-2-(3-(trifluoromethyl)phenyl)isoindoline-4-carboxylic acid (compound 17) | | Descriptor: | 3-oxidanylidene-2-[3-(trifluoromethyl)phenyl]-1~{H}-isoindole-4-carboxylic acid, GLYCEROL, Metallo-beta-lactamase VIM-2, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2016-06-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | NMR-filtered virtual screening leads to non-metal chelating metallo-beta-lactamase inhibitors.
Chem Sci, 8, 2017
|
|
5LE1
 
 | | VIM-2 metallo-beta-lactamase in complex with 2-(2-chloro-6-fluorobenzyl)-3-oxoisoindoline-4-carboxylic acid (compound 16) | | Descriptor: | 2-[(2-chloranyl-6-fluoranyl-phenyl)methyl]-3-oxidanylidene-1~{H}-isoindole-4-carboxylic acid, FORMIC ACID, GLYCEROL, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2016-06-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | NMR-filtered virtual screening leads to non-metal chelating metallo-beta-lactamase inhibitors.
Chem Sci, 8, 2017
|
|
5LM6
 
 | | VIM-2 metallo-beta-lactamase in complex with 2-(3-fluoro-4-hydroxyphenyl)-3-oxoisoindoline-4-carboxylic acid (compound 35) | | Descriptor: | 2-(3-fluoranyl-4-oxidanyl-phenyl)-3-oxidanylidene-1~{H}-isoindole-4-carboxylic acid, FORMIC ACID, Metallo-beta-lactamase VIM-2, ... | | Authors: | Li, G.-B, Brem, J, Someya, H, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2016-07-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | NMR-filtered virtual screening leads to non-metal chelating metallo-beta-lactamase inhibitors.
Chem Sci, 8, 2017
|
|
5LCF
 
 | | VIM-2 metallo-beta-lactamase in complex with 3-oxo-2-phenylisoindoline-4-carboxylic acid (compound 30) | | Descriptor: | 3-oxidanylidene-2-phenyl-1~{H}-isoindole-4-carboxylic acid, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2016-06-21 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | NMR-filtered virtual screening leads to non-metal chelating metallo-beta-lactamase inhibitors.
Chem Sci, 8, 2017
|
|
6HD2
 
 | |
5XBO
 
 | | Lanthanoid tagging via an unnatural amino acid for protein structure characterization | | Descriptor: | Polyubiquitin-B, TERBIUM(III) ION, UV excision repair protein RAD23 homolog A | | Authors: | Jiang, W, Gu, X, Dong, X, Tang, C. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Lanthanoid tagging via an unnatural amino acid for protein structure characterization
J. Biomol. NMR, 67, 2017
|
|
6M0Y
 
 | | KR-12 analog derived from human LL-37 | | Descriptor: | LYS-ARG-ILE-VAL-LYS-ARG-ILE-LYS-LYS-TRP-LEU-ARG | | Authors: | Yun, H, Min, H.J, Lee, C.W. | | Deposit date: | 2020-02-24 | | Release date: | 2021-02-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Bactericidal Activity of KR-12 Analog Derived from Human LL-37 as a Potential Cosmetic Preservative
To Be Published
|
|
