2RDT
 
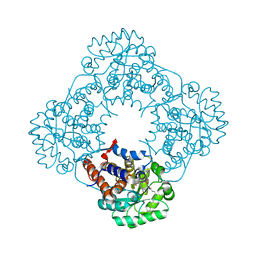 | | Crystal Structure of Human Glycolate Oxidase (GO) in Complex with CDST | | Descriptor: | 5-(dodecylthio)-1H-1,2,3-triazole-4-carboxylic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Murray, M.S, Holmes, R.P, Lowther, W.T. | | Deposit date: | 2007-09-25 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Active Site and Loop 4 Movements within Human Glycolate Oxidase: Implications for Substrate Specificity and Drug Design.
Biochemistry, 47, 2008
|
|
4Q39
 
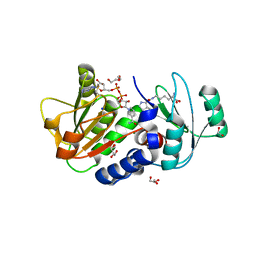 | | PylD in complex with pyrrolysine and NADH | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Quitterer, F, Beck, P, Bacher, A, Groll, M. | | Deposit date: | 2014-04-11 | | Release date: | 2014-04-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Formation of Pyrroline and Tetrahydropyridine Rings in Amino Acids Catalyzed by Pyrrolysine Synthase (PylD).
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3CV6
 
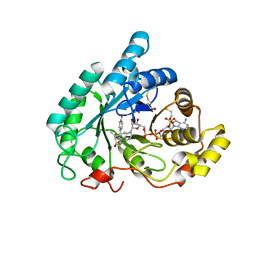 | | The crystal structure of mouse 17-alpha hydroxysteroid dehydrogenase GG225.226PP mutant in complex with inhibitor and cofactor NADP+. | | Descriptor: | 4-[(1R,2S)-1-ethyl-2-(4-hydroxyphenyl)butyl]phenol, Aldo-keto reductase family 1 member C21, BETA-MERCAPTOETHANOL, ... | | Authors: | Dhagat, U, El-Kabbani, O. | | Deposit date: | 2008-04-17 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the G225P/G226P mutant of mouse 3(17)alpha-hydroxysteroid dehydrogenase (AKR1C21) ternary complex: implications for the binding of inhibitor and substrate.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2RBW
 
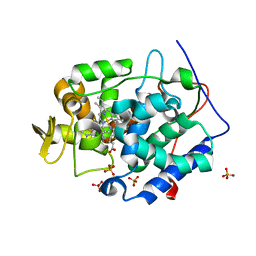 | | Cytochrome C Peroxidase W191G in complex with 1,2-dimethyl-1h-pyridin-5-amine | | Descriptor: | 5-amino-1,2-dimethylpyridinium, Cytochrome C Peroxidase, PHOSPHATE ION, ... | | Authors: | Graves, A.P, Boyce, S.E, Shoichet, B.K. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rescoring docking hit lists for model cavity sites: predictions and experimental testing.
J.Mol.Biol., 377, 2008
|
|
4NBV
 
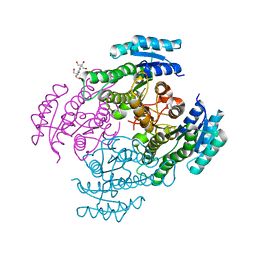 | | Crystal structure of FabG from Cupriavidus taiwanensis | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-oxoacyl-[acyl-carrier-protein] reductase putative short-chain dehydrogenases/reductases (SDR) family protein | | Authors: | Pereira, J.H, Mcandrew, R.P, Javidpour, P, Beller, H.R, Adams, P.D. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.645 Å) | | Cite: | Biochemical and Structural Studies of NADH-Dependent FabG Used To Increase the Bacterial Production of Fatty Acids under Anaerobic Conditions.
Appl.Environ.Microbiol., 80, 2014
|
|
1BU7
 
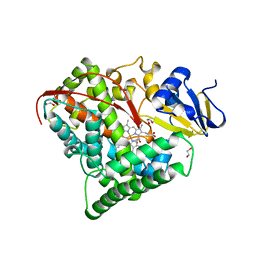 | | CRYOGENIC STRUCTURE OF CYTOCHROME P450BM-3 HEME DOMAIN | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN (CYTOCHROME P450), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 1998-09-14 | | Release date: | 1998-09-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of a cytochrome P450-redox partner electron-transfer complex.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1T7Q
 
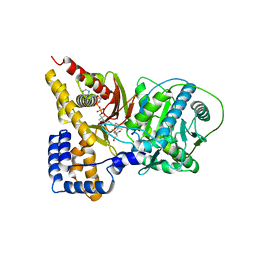 | | Crystal structure of the F565A mutant of murine carnitine acetyltransferase in complex with carnitine and CoA | | Descriptor: | 1,2-ETHANEDIOL, CARNITINE, COENZYME A, ... | | Authors: | Hsiao, Y.-S, Jogl, G, Tong, L. | | Deposit date: | 2004-05-10 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical studies of the substrate selectivity of carnitine acetyltransferase
J.Biol.Chem., 279, 2004
|
|
5AC2
 
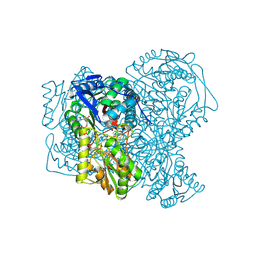 | | human aldehyde dehydrogenase 1A1 with duocarmycin analog | | Descriptor: | 1-[(1S)-1-methyl-5-oxidanyl-1,2-dihydrobenzo[e]indol-3-yl]hexan-1-one, RETINAL DEHYDROGENASE 1, YTTERBIUM (III) ION, ... | | Authors: | Koch, M.F, Harteis, S, Blank, I.D, Pestel, G, Tietze, L.F, Ochsenfeld, C, Schneider, S, Sieber, S.A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural, Biochemical, and Computational Studies Reveal the Mechanism of Selective Aldehyde Dehydrogenase 1A1 Inhibition by Cytotoxic Duocarmycin Analogues.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4IIU
 
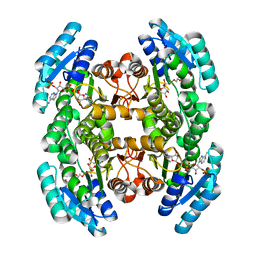 | | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Escherichia coli strain CFT073 complexed with NADP+ at 2.1 A resolution | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hou, J, Osinski, T, Zheng, H, Shumilin, I, Shabalin, I, Shatsman, S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-20 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Escherichia coli strain CFT073 complexed with NADP+ at 2.1 A resolution
To be Published
|
|
3RPE
 
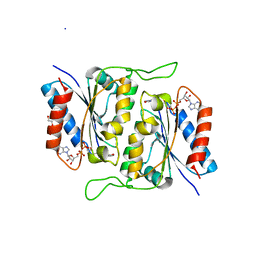 | | 1.1 Angstrom Crystal Structure of Putative Modulator of Drug Activity (MdaB) from Yersinia pestis CO92. | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, Modulator of drug activity B | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-26 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | 1.1 Angstrom Crystal Structure of Putative Modulator of Drug Activity (MdaB) from Yersinia pestis CO92.
TO BE PUBLISHED
|
|
7F4V
 
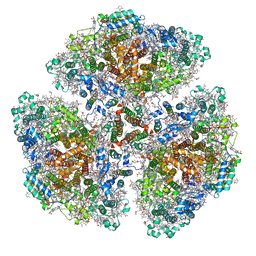 | | Cryo-EM structure of a primordial cyanobacterial photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Hamaguchi, T, Nagao, R, Kawakami, K, Yonekura, K, Shen, J.R. | | Deposit date: | 2021-06-21 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.04 Å) | | Cite: | Structural basis for the absence of low-energy chlorophylls responsible for photoprotection from a primitive cyanobacterial PSI
Biorxiv, 2022
|
|
5DKG
 
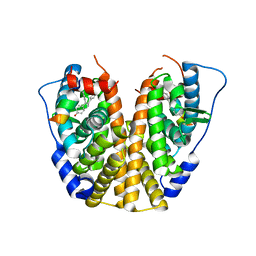 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a t-butyl-substituted, methyl, triaryl-ethylene derivative 4,4'-[2-(4-tert-butylphenyl)prop-1-ene-1,1-diyl]diphenol | | Descriptor: | 4,4'-[2-(4-tert-butylphenyl)prop-1-ene-1,1-diyl]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-03 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
2BL7
 
 | | 1.6 Angstrom crystal structure of EntA-im: a bacterial immunity protein conferring immunity to the antimicrobial activity of the pediocin-like bacteriocin, enterocin A | | Descriptor: | ENTEROCINE A IMMUNITY PROTEIN | | Authors: | Johnsen, L, Dalhus, B, Leiros, I, Nissen-Meyer, J. | | Deposit date: | 2005-03-02 | | Release date: | 2005-03-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 1.6-A Crystal Structure of Enta-Im: A Bacterial Immunity Protein Conferring Immunity to the Antimicrobial Activity of the Pediocin-Like Bacteriocin Enterocin A
J.Biol.Chem., 280, 2005
|
|
3N8M
 
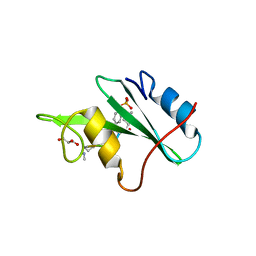 | |
3I1J
 
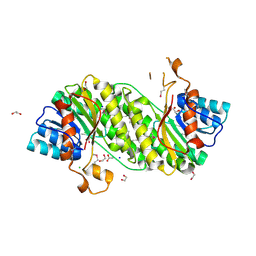 | | Structure of a putative short chain dehydrogenase from Pseudomonas syringae | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Singer, A.U, Evdokimova, E, Kudritska, M, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-06-26 | | Release date: | 2009-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a putative short chain dehydrogenase from Pseudomonas syringae
To be Published
|
|
5E70
 
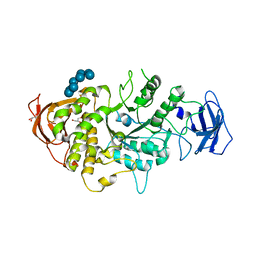 | | Crystal structure of Ecoli Branching Enzyme with gamma cyclodextrin | | Descriptor: | 1,4-alpha-glucan branching enzyme GlgB, Cyclooctakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL | | Authors: | Feng, L, Nosrati, M, Geiger, J.H. | | Deposit date: | 2015-10-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structures of Escherichia coli branching enzyme in complex with cyclodextrins.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5E6Z
 
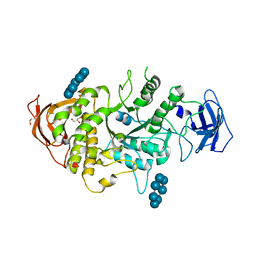 | | Crystal structure of Ecoli Branching Enzyme with beta cyclodextrin | | Descriptor: | 1,4-alpha-glucan branching enzyme GlgB, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL | | Authors: | Feng, L, Nosrati, M, Geiger, J.H. | | Deposit date: | 2015-10-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Crystal structures of Escherichia coli branching enzyme in complex with cyclodextrins.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2BL8
 
 | | 1.6 Angstrom crystal structure of EntA-im: a bacterial immunity protein conferring immunity to the antimicrobial activity of the pediocin-like bacteriocin, enterocin A | | Descriptor: | CITRATE ANION, ENTEROCINE A IMMUNITY PROTEIN | | Authors: | Johnsen, L, Dalhus, B, Leiros, I, Nissen-Meyer, J. | | Deposit date: | 2005-03-02 | | Release date: | 2005-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.6-A Crystal Structure of Enta-Im: A Bacterial Immunity Protein Conferring Immunity to the Antimicrobial Activity of the Pediocin-Like Bacteriocin Enterocin A
J.Biol.Chem., 280, 2005
|
|
1O76
 
 | | CYANIDE COMPLEX OF P450CAM FROM PSEUDOMONAS PUTIDA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYANIDE ION, ... | | Authors: | Fedorov, R, Ghosh, D, Schlichting, I. | | Deposit date: | 2002-10-23 | | Release date: | 2002-12-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Cyanide Complexes of P450Cam and the Oxygenase Domain of Inducible Nitric Oxide Synthase-Structural Models of the Short-Lived Oxygen Complexes
Arch.Biochem.Biophys., 409, 2003
|
|
6EV2
 
 | | Crystal structure of antibody against schizophyllan in complex with laminarihexaose | | Descriptor: | Heavy chain, Light chain, beta-D-glucopyranose, ... | | Authors: | Sung, K.H, Josewski, J, Duebel, S, Blankenfeldt, W, Rau, U. | | Deposit date: | 2017-11-01 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Structural insights into antigen recognition of an anti-beta-(1,6)-beta-(1,3)-D-glucan antibody.
Sci Rep, 8, 2018
|
|
5E6Y
 
 | | Crystal structure of E.Coli branching enzyme in complex with alpha cyclodextrin | | Descriptor: | 1,4-alpha-glucan branching enzyme GlgB, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL | | Authors: | Feng, L, Nosrati, M, Geiger, J.H. | | Deposit date: | 2015-10-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of Escherichia coli branching enzyme in complex with cyclodextrins.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1OAA
 
 | | MOUSE SEPIAPTERIN REDUCTASE COMPLEXED WITH NADP AND OXALOACETATE | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OXALOACETATE ION, SEPIAPTERIN REDUCTASE, ... | | Authors: | Auerbach, G, Herrmann, A, Bacher, A, Huber, R. | | Deposit date: | 1997-08-25 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The 1.25 A crystal structure of sepiapterin reductase reveals its binding mode to pterins and brain neurotransmitters.
EMBO J., 16, 1997
|
|
1AMM
 
 | | 1.2 ANGSTROM STRUCTURE OF GAMMA-B CRYSTALLIN AT 150K | | Descriptor: | GAMMA B-CRYSTALLIN | | Authors: | Kumaraswamy, V.S, Lindley, P.F, Slingsby, C, Glover, I.D. | | Deposit date: | 1996-03-20 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An eye lens protein-water structure: 1.2 A resolution structure of gammaB-crystallin at 150 K.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
4AP9
 
 | | Crystal structure of phosphoserine phosphatase from T. onnurineus in complex with NDSB-201 | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, PHOSPHOSERINE PHOSPHATASE | | Authors: | Jung, T.-Y, Kim, Y.-S, Song, H.-N, Woo, E. | | Deposit date: | 2012-03-31 | | Release date: | 2012-12-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Identification of a Novel Ligand Binding Site in Phosphoserine Phosphatase from the Hyperthermophilic Archaeon Thermococcus Onnurineus.
Proteins, 81, 2013
|
|
5T0V
 
 | | Architecture of the Yeast Mitochondrial Iron-Sulfur Cluster Assembly Machinery: the Sub-Complex Formed by the Iron Donor, Yfh1, and the Scaffold, Isu1 | | Descriptor: | Frataxin homolog, mitochondrial, Iron sulfur cluster assembly protein 1 | | Authors: | Ranatunga, W, Gakh, O, Galeano, B.K, Smith IV, D.Y, Soderberg, C.A, Al-Karadaghi, S, Thompson, J.R, Isaya, G. | | Deposit date: | 2016-08-16 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (17.5 Å) | | Cite: | Architecture of the Yeast Mitochondrial Iron-Sulfur Cluster Assembly Machinery: the Sub-Complex Formed by the Iron Donor, Yfh1, and the Scaffold, Isu1
J. Biol. Chem., 291, 2016
|
|
