6TJD
 
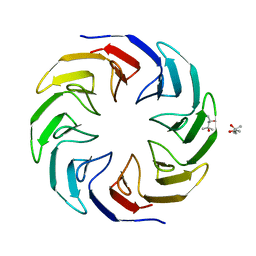 | | Crystal structure of the computationally designed Cake4 protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cake4 | | Authors: | Laier, I, Mylemans, B, Noguchi, H, Voet, A.R.D. | | Deposit date: | 2019-11-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
5OK3
 
 | | Crystal Structure of the Protein-Kinase A catalytic subunit from Criteculus Griseus in complex with compounds RKp241 and Fasudil | | Descriptor: | 5-(1,4-DIAZEPAN-1-SULFONYL)ISOQUINOLINE, UPF0418 protein FAM164A, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Mueller, J.M, Heine, A, Klebe, G. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.588 Å) | | Cite: | Diamondoid Amino Acid-Based Peptide Kinase A Inhibitor Analogues.
Chemmedchem, 14, 2019
|
|
3F7P
 
 | |
5UYN
 
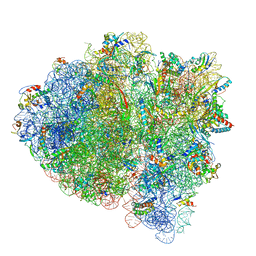 | | 70S ribosome bound with near-cognate ternary complex not base-paired to A site codon (Structure I-nc) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity
Nature, 546, 2017
|
|
5HA7
 
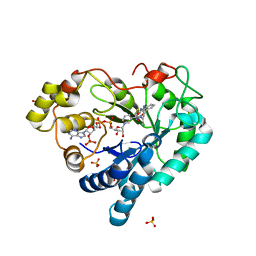 | | Human Aldose Reductase in Complex with NADP+ and WY14643 in Space Group P212121 | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sawaya, M.R, Cascio, D, Balendiran, G.K. | | Deposit date: | 2015-12-30 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Characterization of WY 14,643 and its Complex with Aldose Reductase.
Sci Rep, 6, 2016
|
|
3ENR
 
 | | ZINC-CALCIUM CONCANAVALIN A AT PH 6.15 | | Descriptor: | CALCIUM ION, Concanavalin-A, ZINC ION | | Authors: | Bouckaert, J, Loris, R, Wyns, L. | | Deposit date: | 1999-02-11 | | Release date: | 2000-09-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Zinc/calcium- and cadmium/cadmium-substituted concanavalin A: interplay of metal binding, pH and molecular packing.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
8P5R
 
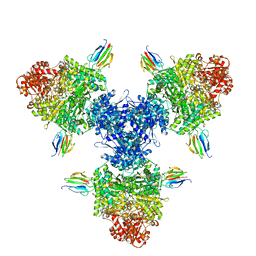 | | Crystal structure of full-length, homohexameric 2-oxoglutarate dehydrogenase KGD from Mycobacterium smegmatis in complex with GarA | | Descriptor: | CALCIUM ION, Glycogen accumulation regulator GarA, MAGNESIUM ION, ... | | Authors: | Wagner, T, Mechaly, A.M, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.562 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
1PE4
 
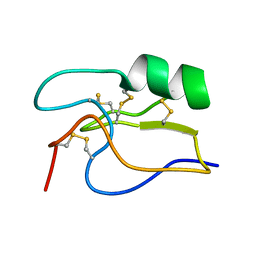 | | SOLUTION STRUCTURE OF TOXIN CN12 FROM CENTRUROIDES NOXIUS ALFA SCORPION TOXIN ACTING ON SODIUM CHANNELS. NMR STRUCTURE | | Descriptor: | Neurotoxin Cn11 | | Authors: | Del Rio-Portilla, F, Hernandez-Marin, E, Pimienta, G, Coronas, F.V, Zamudio, F.Z, Rodrguez de la Vega, R.C, Wanke, E, Possani, L.D. | | Deposit date: | 2003-05-21 | | Release date: | 2004-05-25 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of Cn12, a novel peptide from the Mexican scorpion Centruroides noxius with a typical beta-toxin sequence but with alpha-like physiological activity.
Eur.J.Biochem., 271, 2004
|
|
5III
 
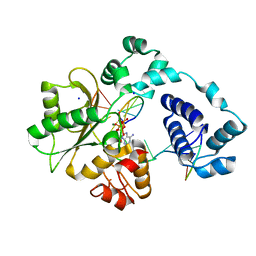 | | Crystal structure of the pre-catalytic ternary complex of DNA polymerase lambda with a templating 8-oxo-dG and an incoming dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*AP*(DOC))-3'), DNA (5'-D(*CP*GP*GP*CP*(8OG)P*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Burak, M.J, Guja, K.E, Garcia-Diaz, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A fidelity mechanism in DNA polymerase lambda promotes error-free bypass of 8-oxo-dG.
Embo J., 35, 2016
|
|
1LHM
 
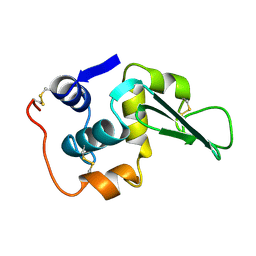 | |
1L58
 
 | |
1L64
 
 | |
1L71
 
 | |
1L76
 
 | |
5TWP
 
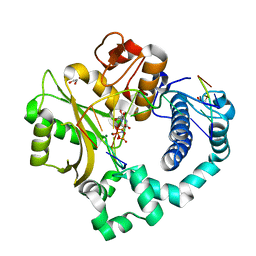 | | Pre-catalytic ternary complex of human Polymerase Mu with incoming nonhydrolyzable UMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, DNA (5'-D(*CP*GP*GP*CP*AP*TP*AP*CP*G)-3'), ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2016-11-14 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
1OBF
 
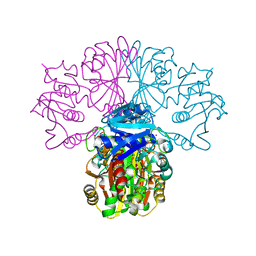 | | The crystal structure of Glyceraldehyde 3-phosphate Dehydrogenase from Alcaligenes xylosoxidans at 1.7A resolution. | | Descriptor: | GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE, POTASSIUM ION, SULFATE ION, ... | | Authors: | Antonyuk, S.V, Eady, R.R, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Glyceraldehyde 3-Phosphate Dehydrogenase from Alcaligenes Xylosoxidans at 1.7 A Resolution
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1ODK
 
 | |
5MVT
 
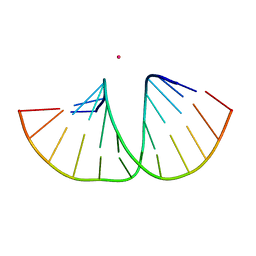 | | Crystal structure of an A-DNA dodecamer featuring an alternating pyrimidine-purine sequence | | Descriptor: | COBALT (III) ION, DNA | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5VZG
 
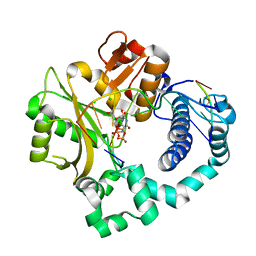 | | Pre-catalytic ternary complex of human Polymerase Mu (W434H) mutant with incoming nonhydrolyzable UMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, CHLORIDE ION, ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2017-05-27 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
2LQL
 
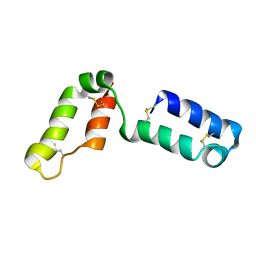 | | Solution structure of CHCH5 | | Descriptor: | Coiled-coil-helix-coiled-coil-helix domain-containing protein 5 | | Authors: | Peruzzini, R, Ciofi-Baffoni, S, Banci, L, Bertini, I. | | Deposit date: | 2012-03-09 | | Release date: | 2012-10-24 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of CHCHD5 and CHCHD7: Two atypical human twin CX(9)C proteins.
J.Struct.Biol., 180, 2012
|
|
3DRU
 
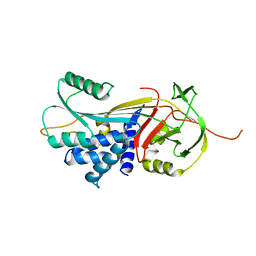 | | Crystal Structure of Gly117Phe Alpha1-Antitrypsin | | Descriptor: | Alpha-1-antitrypsin | | Authors: | Gooptu, B, Nobeli, I, Purkiss, A, Phillips, R.L, Mallya, M, Lomas, D.A, Barrett, T.E. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystallographic and cellular characterisation of two mechanisms stabilising the native fold of alpha1-antitrypsin: implications for disease and drug design.
J.Mol.Biol., 387, 2009
|
|
6T6H
 
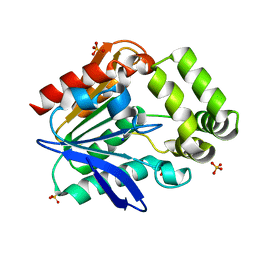 | | Apo structure of the Bottromycin epimerase BotH | | Descriptor: | BotH, SODIUM ION, SULFATE ION | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2019-10-18 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | The bottromycin epimerase BotH defines a group of atypical alpha / beta-hydrolase-fold enzymes.
Nat.Chem.Biol., 16, 2020
|
|
1PUA
 
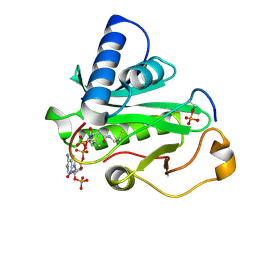 | | Crystal Structure of Tetrahymena GCN5 with Bound Coenzyme A and a Phosphorylated, 19-residue Histone H3 peptide | | Descriptor: | COENZYME A, HAT A1, Histone H3 | | Authors: | Clements, A, Poux, A.N, Lo, W.S, Pillus, L, Berger, S.L, Marmorstein, R. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for histone and phospho-histone binding by the GCN5 histone acetyltransferase
Mol.Cell, 12, 2003
|
|
6T6Z
 
 | |
1PS2
 
 | | HIGH RESOLUTION NMR SOLUTION STRUCTURE OF HUMAN PS2, 19 STRUCTURES | | Descriptor: | PS2 | | Authors: | Williams, M.A, Polshakov, V.I, Gargaro, A.R, Feeney, J. | | Deposit date: | 1997-01-07 | | Release date: | 1997-07-07 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of human pNR-2/pS2: a single trefoil motif protein.
J.Mol.Biol., 267, 1997
|
|
