3JWF
 
 | | Crystal structure of Bacillus anthracis (Y102F) dihydrofolate reductase complexed with NADPH and (R)-2,4-diamino-5-(3-hydroxy-3-(3,4,5-trimethoxyphenyl)prop-1-ynyl)-6-methylpyrimidine (UCP113A) | | Descriptor: | (1R)-3-(2,4-diamino-6-methylpyrimidin-5-yl)-1-(3,4,5-trimethoxyphenyl)prop-2-yn-1-ol, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Anderson, A.C, Beierlein, J.M, Karri, N.G. | | Deposit date: | 2009-09-18 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Targeted Mutations in Bacillus anthracis Dihydrofolate Reductase Condense Complex Structure-Activity Relationships
J.Med.Chem., 2010
|
|
3H3F
 
 | | Rabbit muscle L-lactate dehydrogenase in complex with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETATE ION, L-lactate dehydrogenase A chain, ... | | Authors: | Bujacz, A, Bujacz, G, Swiderek, K, Paneth, P. | | Deposit date: | 2009-04-16 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Modeling of isotope effects on binding oxamate to lactic dehydrogenase
J.Phys.Chem.B, 113, 2009
|
|
5ZE1
 
 | | Hairpin Forming Complex, RAG1/2-Nicked 12RSS/23RSS complex in 2mM Mn2+ for 10 min at 4'C | | Descriptor: | 1,2-ETHANEDIOL, DNA, HMGB1 A-B box, ... | | Authors: | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2018-02-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
6M6L
 
 | | The crystal structure of glycosidase hydrolyzing Notoginsenoside | | Descriptor: | 1,2-ETHANEDIOL, Beta-glucosidase, GLYCEROL, ... | | Authors: | Wang, R.F. | | Deposit date: | 2020-03-15 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization and Structural Elucidation of Enhanced Catalytic Activity upon Engineering Glycosidase KfGH01 for the Production of Vina-ginsenoside R7
To Be Published
|
|
3JWM
 
 | | Crystal structure of Bacillus anthracis dihydrofolate reductase complexed with NADPH and (S)-2,4-diamino-5-(3-methoxy-3-(3,4,5-trimethoxyphenyl)prop-1-ynyl)-6-methylpyrimidine (UCP114A) | | Descriptor: | 5-[(3S)-3-methoxy-3-(3,4,5-trimethoxyphenyl)prop-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Anderson, A.C, Beierlein, J.M, Karri, N.G. | | Deposit date: | 2009-09-18 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Mutational Studies into Trimethoprim Resistance in Bacillus anthracis Dihydrofolate Reductase
To be Published
|
|
4N11
 
 | |
4N10
 
 | |
1Z6N
 
 | | 1.5 A Crystal Structure of a Protein of Unknown Function PA1234 from Pseudomonas aeruginosa | | Descriptor: | MAGNESIUM ION, hypothetical protein PA1234 | | Authors: | Zhang, R, Xu, L, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-22 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein PA1234 from Pseudomonas aeruginosa
To be Published
|
|
5VBC
 
 | | Crystal structure of ATXR5 in complex with histone H3.1 | | Descriptor: | DIMETHYL SULFOXIDE, Histone H3.1 peptide, Probable Histone-lysine N-methyltransferase ATXR5, ... | | Authors: | Couture, J.-F, Bergamin, E. | | Deposit date: | 2017-03-29 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
6M2U
 
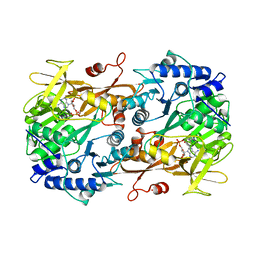 | |
5OV6
 
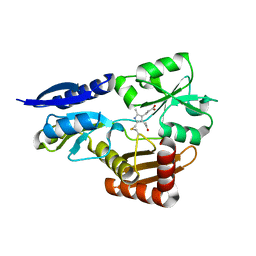 | | Bacillus megaterium porphobilinogen deaminase D82N mutant | | Descriptor: | 3-[4-(2-hydroxy-2-oxoethyl)-2,5-dimethyl-1~{H}-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Guo, J, Erskine, P, Coker, A.R, Wood, S.P, Cooper, J.B. | | Deposit date: | 2017-08-28 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural studies of domain movement in active-site mutants of porphobilinogen deaminase from Bacillus megaterium.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
1CTL
 
 | | STRUCTURE OF THE CARBOXY-TERMINAL LIM DOMAIN FROM THE CYSTEINE RICH PROTEIN CRP | | Descriptor: | AVIAN CYSTEINE RICH PROTEIN, ZINC ION | | Authors: | Perez-Alvarado, G.C, Miles, C, Michelsen, J.W, Louis, H.A, Winge, D.R, Beckerle, M.C, Summers, M.F. | | Deposit date: | 1995-01-06 | | Release date: | 1995-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the carboxy-terminal LIM domain from the cysteine rich protein CRP.
Nat.Struct.Biol., 1, 1994
|
|
3CLB
 
 | | Structure of bifunctional TcDHFR-TS in complex with TMQ | | Descriptor: | 1,2-ETHANEDIOL, DHFR-TS, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schormann, N, Senkovich, O, Chattopadhyay, D. | | Deposit date: | 2008-03-18 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based approach to pharmacophore identification, in silico screening, and three-dimensional quantitative structure-activity relationship studies for inhibitors of Trypanosoma cruzi dihydrofolate reductase function.
Proteins, 73, 2008
|
|
2WDZ
 
 | | Crystal structure of the short chain dehydrogenase Galactitol- Dehydrogenase (GatDH) of Rhodobacter sphaeroides in complex with NAD+ and 1,2-Pentandiol | | Descriptor: | (2S)-pentane-1,2-diol, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Carius, Y, Christian, H, Faust, A, Kornberger, P, Kohring, G.W, Giffhorn, F, Scheidig, A.J. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insight Into Substrate Differentiation of the Sugar-Metabolizing Enzyme Galactitol Dehydrogenase from Rhodobacter Sphaeroides D.
J.Biol.Chem., 285, 2010
|
|
4QX6
 
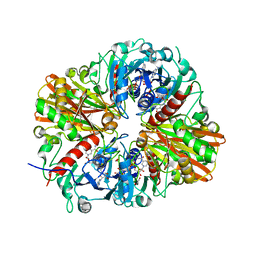 | | CRYSTAL STRUCTURE OF GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE FROM STREPTOCOCCUS AGALACTIAE NEM316 at 2.46 ANGSTROM RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Ayres, C.A, Schormann, N, Banerjee, S, Chattopadhyay, D. | | Deposit date: | 2014-07-18 | | Release date: | 2014-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure of Streptococcus agalactiae glyceraldehyde-3-phosphate dehydrogenase holoenzyme reveals a novel surface.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
5ZE2
 
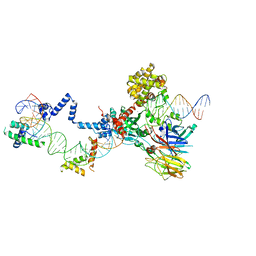 | | Hairpin Complex, RAG1/2-hairpin 12RSS/23RSS complex in 5mM Mn2+ for 2 min at 4'C | | Descriptor: | 1,2-ETHANEDIOL, DNA (30-MER), DNA (31-MER), ... | | Authors: | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2018-02-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
6K9S
 
 | | Structure of the Carbonylruthenium Mesoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | Descriptor: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, CARBON MONOXIDE, ... | | Authors: | Stanfield, J.K, Omura, K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-06-17 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6KMW
 
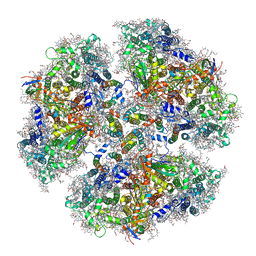 | | Structure of PSI from H. hongdechloris grown under white light condition | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-08-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structural basis for the adaptation and function of chlorophyll f in photosystem I.
Nat Commun, 11, 2020
|
|
1ARZ
 
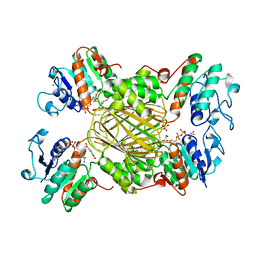 | | ESCHERICHIA COLI DIHYDRODIPICOLINATE REDUCTASE IN COMPLEX WITH NADH AND 2,6 PYRIDINE DICARBOXYLATE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIHYDRODIPICOLINATE REDUCTASE, PHOSPHATE ION, ... | | Authors: | Scapin, G, Reddy, S.G, Zheng, R, Blanchard, J.S. | | Deposit date: | 1997-08-08 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structure of Escherichia coli dihydrodipicolinate reductase in complex with NADH and the inhibitor 2,6-pyridinedicarboxylate.
Biochemistry, 36, 1997
|
|
7JZ2
 
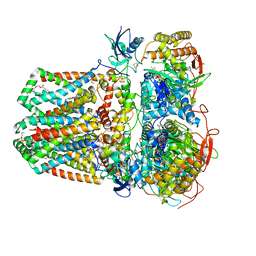 | | Succinate: quinone oxidoreductase SQR from E.coli K12 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Lyu, M, Su, C.-C, Morgan, C.E, Bolla, J.R, Robinson, C.V, Yu, E.W. | | Deposit date: | 2020-09-01 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
5V4R
 
 | |
7KLI
 
 | |
6MJT
 
 | | Azurin 122F/124W/126Re | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (II) ION | | Authors: | Takematsu, K, Zalis, S, Gray, H.B, Vlcek, A, Winkler, J.R, Williamson, H, Kaiser, J.T, Heyda, J, Hollas, D. | | Deposit date: | 2018-09-21 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Two Tryptophans Are Better Than One in Accelerating Electron Flow through a Protein.
ACS Cent Sci, 5, 2019
|
|
6MJR
 
 | | Azurin 122W/124F/126Re | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (II) ION | | Authors: | Takematsu, K, Zalis, S, Gray, H.B, Vlcek, A, Winkler, J.R, Williamson, H, Kaiser, J.T, Heyda, J, Hollas, D. | | Deposit date: | 2018-09-21 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Two Tryptophans Are Better Than One in Accelerating Electron Flow through a Protein.
ACS Cent Sci, 5, 2019
|
|
4PVL
 
 | | X-ray structure of human transthyretin (TTR) at room temperature to 1.9A resolution | | Descriptor: | Transthyretin | | Authors: | Fisher, S.J, Blakeley, M.P, Haupt, M, Mason, S.A, Cooper, J.B, Mitchell, E.P, Forsyth, V.T. | | Deposit date: | 2014-03-18 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Binding site asymmetry in human transthyretin: insights from a joint neutron and X-ray crystallographic analysis using perdeuterated protein
IUCrJ, 1, 2014
|
|
