5LUP
 
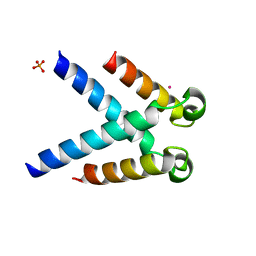 | | Structures of DHBN domain of human BLM helicase | | Descriptor: | BLM protein, PHOSPHATE ION, POTASSIUM ION | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
8T02
 
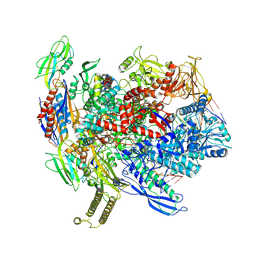 | | Reconstituted E. coli RNA polymerase post-termination complex on negatively-supercoiled DNA: unwinding duplex DNA (rPTCi) | | Descriptor: | DNA (25-MER), DNA (26-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Brewer, J.J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-31 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | RapA opens the RNA polymerase clamp to disrupt post-termination complexes and prevent cytotoxic R-loop formation.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8SZW
 
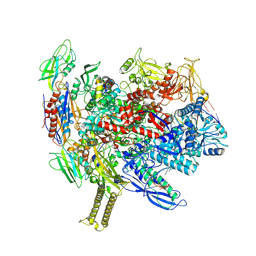 | | Reconstituted E. coli RNA polymerase post-termination complex on negatively-supercoiled DNA: open duplex DNA (rPTCo) | | Descriptor: | DNA (25-MER), DNA (27-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Brewer, J.J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-30 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | RapA opens the RNA polymerase clamp to disrupt post-termination complexes and prevent cytotoxic R-loop formation.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8T00
 
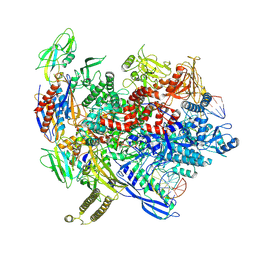 | | Reconstituted E. coli RNA polymerase post-termination complex on negatively-supercoiled DNA: closed duplex DNA (rPTCc) | | Descriptor: | DNA (26-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Brewer, J.J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-31 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | RapA opens the RNA polymerase clamp to disrupt post-termination complexes and prevent cytotoxic R-loop formation.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8T0L
 
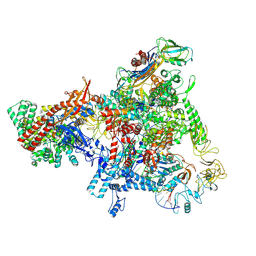 | | E. coli Sw2/Snf2 ATPase RapA bound to both ADP-AlF3 and reconstituted E. coli RNA polymerase post-termination complex on negatively-supercoiled DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, DNA (29-MER), ... | | Authors: | Brewer, J.J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-06-01 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | RapA opens the RNA polymerase clamp to disrupt post-termination complexes and prevent cytotoxic R-loop formation.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6NWX
 
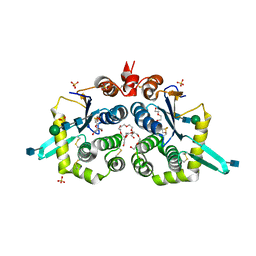 | | Structure of mouse GILT, an enzyme involved in antigen processing | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-interferon-inducible lysosomal thiol reductase, PENTAETHYLENE GLYCOL, ... | | Authors: | Li, Y, Cresswell, P. | | Deposit date: | 2019-02-07 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Gamma-interferon-inducible lysosomal thiol reductase (GILT). Maturation, activity, and mechanism of action
To Be Published
|
|
5LUT
 
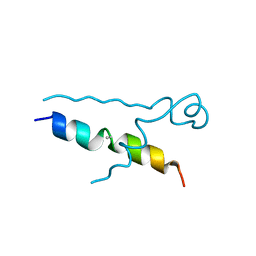 | | Structures of DHBN domain of Gallus gallus BLM helicase | | Descriptor: | BLM helicase, PHOSPHATE ION | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
9B68
 
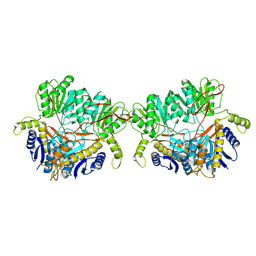 | | GluA2 flip Q in complex with TARPgamma2 at pH8, class1, structure of NTD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Flip of Glutamate receptor 2, ... | | Authors: | Nakagawa, T, Greger, I.H. | | Deposit date: | 2024-03-23 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Proton-triggered rearrangement of the AMPA receptor N-terminal domains impacts receptor kinetics and synaptic localization.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6OJK
 
 | | Structure of YePL2A K291W in Complex with Tetragalacturonic Acid | | Descriptor: | 1,2-ETHANEDIOL, Periplasmic pectate lyase, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Jones, D.R, Abbott, D.W. | | Deposit date: | 2019-04-11 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A surrogate structural platform informed by ancestral reconstruction and resurrection of a putative carbohydrate binding module hybrid illuminates the neofunctionalization of a pectate lyase.
J.Struct.Biol., 207, 2019
|
|
6IM8
 
 | | CueO-PM2 multicopper oxidase | | Descriptor: | Blue copper oxidase CueO,PM2 peptide,Blue copper oxidase CueO | | Authors: | Wongsantichon, J, Robinson, R, Ghadessy, F. | | Deposit date: | 2018-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Development and structural characterization of an engineered multi-copper oxidase reporter of protein-protein interactions.
J.Biol.Chem., 294, 2019
|
|
5LYY
 
 | | Fragment-based inhibitors of Lipoprotein associated Phospholipase A2 | | Descriptor: | 3-[2-(4-fluoranylphenoxy)ethyl]-1,3-diazaspiro[4.5]decane-2,4-dione, Platelet-activating factor acetylhydrolase | | Authors: | Woolford, A, Day, P. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fragment-Based Approach to the Development of an Orally Bioavailable Lactam Inhibitor of Lipoprotein-Associated Phospholipase A2 (Lp-PLA2).
J. Med. Chem., 59, 2016
|
|
5M0E
 
 | | Structure-based evolution of a hybrid steroid series of Autotaxin inhibitors | | Descriptor: | 7alpha-hydroxycholesterol, CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Keune, W.-J, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2016-10-04 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational Design of Autotaxin Inhibitors by Structural Evolution of Endogenous Modulators.
J. Med. Chem., 60, 2017
|
|
8UM1
 
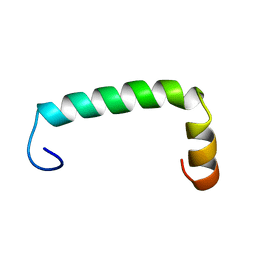 | |
5M0P
 
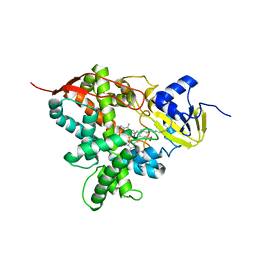 | | Crystal structure of cytochrome P450 OleT F79A in complex with arachidonic acid | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, Terminal olefin-forming fatty acid decarboxylase, ... | | Authors: | Tee, K.L, Munro, A, Matthews, S, Leys, D, Levy, C. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Catalytic Determinants of Alkene Production by the Cytochrome P450 Peroxygenase OleTJE.
J. Biol. Chem., 292, 2017
|
|
5M0X
 
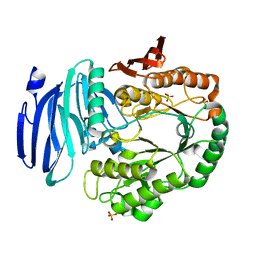 | |
8U4F
 
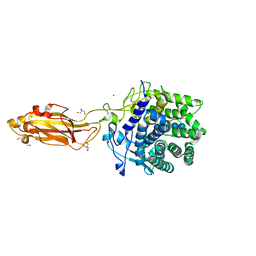 | |
8U4A
 
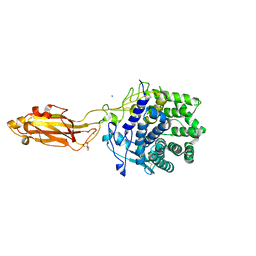 | |
8U49
 
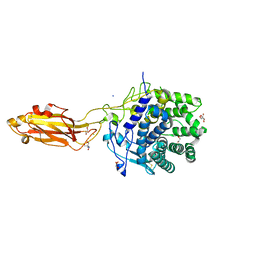 | | The Apo Crystal Structure of BlCel9A from Glycoside Hydrolase Family 9 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Araujo, E.A, Polikarpov, I. | | Deposit date: | 2023-09-10 | | Release date: | 2024-02-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of cellulose depolymerization by the two-domain BlCel9A enzyme from the glycoside hydrolase family 9.
Carbohydr Polym, 329, 2024
|
|
8UM2
 
 | |
7NHP
 
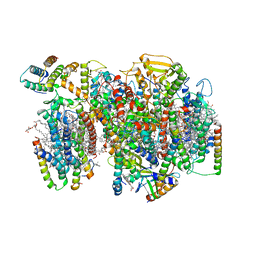 | | Structure of PSII-I (PSII with Psb27, Psb28, and Psb34) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Zabret, J, Bohn, S, Schuller, S.K, Arnolds, O, Chan, A, Tajkhorshid, E, Stoll, R, Engel, B.D, Rudack, T, Schuller, J.M, Nowaczyk, M.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural insights into photosystem II assembly.
Nat.Plants, 7, 2021
|
|
7NHO
 
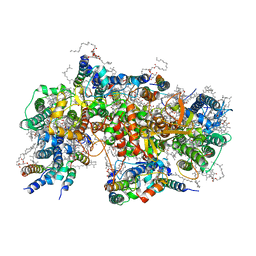 | | Structure of PSII-M | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Zabret, J, Bohn, S, Schuller, S.K, Arnolds, O, Chan, A, Tajkhorshid, E, Stoll, R, Engel, B.D, Rudack, T, Schuller, J.M, Nowaczyk, M.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural insights into photosystem II assembly.
Nat.Plants, 7, 2021
|
|
8UEU
 
 | | In-situ complex I, Deactive class03 | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Zheng, W, Zhu, J, Zhang, K. | | Deposit date: | 2023-10-02 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 631, 2024
|
|
7NHQ
 
 | | Structure of PSII-I prime (PSII with Psb28, and Psb34) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Zabret, J, Bohn, S, Schuller, S.K, Arnolds, O, Chan, A, Tajkhorshid, E, Stoll, R, Engel, B.D, Rudack, T, Schuller, J.M, Nowaczyk, M.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural insights into photosystem II assembly.
Nat.Plants, 7, 2021
|
|
8U7F
 
 | | Crystal structure of CIB_12 beta-galactosidase from Cuniculiplasma divulgatum | | Descriptor: | CIB_12 Beta-galactosidase, GLYCEROL | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Yakunin, A, Golyshin, P, Savchenko, A. | | Deposit date: | 2023-09-15 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Moderately thermostable GH1 beta-glucosidases from hyperacidophilic archaeon Cuniculiplasma divulgatum S5.
Fems Microbiol.Ecol., 100, 2024
|
|
6O4G
 
 | |
