1YI0
 
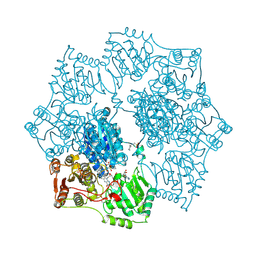 | | Crystal structure of Arabidopsis thaliana Acetohydroxyacid synthase In Complex With A Sulfonylurea Herbicide, Sulfometuron methyl | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Acetolactate synthase, ETHYL DIHYDROGEN DIPHOSPHATE, ... | | Authors: | McCourt, J.A, Pang, S.S, King-Scott, J, Guddat, L.W, Duggleby, R.G. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Herbicide-binding sites revealed in the structure of plant acetohydroxyacid synthase
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6I2T
 
 | | CryoEM reconstruction of full-length, fully-glycosylated human butyrylcholinesterase tetramer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cholinesterase, lamellipodin-derived polyproline peptide | | Authors: | Leung, M.R, van Bezouwen, L.S, Schopfer, L.M, Sussman, J.L, Silman, I, Lockridge, O, Zeev-Ben-Mordehai, T. | | Deposit date: | 2018-11-01 | | Release date: | 2018-12-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Cryo-EM structure of the native butyrylcholinesterase tetramer reveals a dimer of dimers stabilized by a superhelical assembly.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3MEG
 
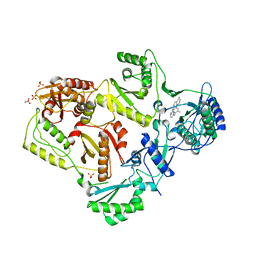 | | HIV-1 K103N Reverse Transcriptase in Complex with TMC278 | | Descriptor: | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, SULFATE ION, p51 Reverse transcriptase, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase with Etravirine (TMC125) and Rilpivirine (TMC278): Implications for Drug Design.
J.Med.Chem., 53, 2010
|
|
1YIU
 
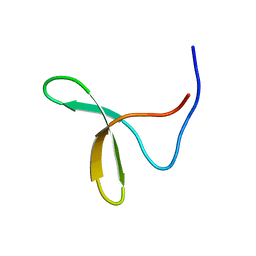 | | Itch E3 ubiquitin ligase WW3 domain | | Descriptor: | Itchy E3 ubiquitin protein ligase | | Authors: | Shaw, A.Z, Martin-Malpartida, P, Morales, B, Yraola, F, Royo, M, Macias, M.J. | | Deposit date: | 2005-01-13 | | Release date: | 2005-08-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation of either Ser16 or Thr30 does not disrupt the structure of the Itch E3 ubiquitin ligase third WW domain
Proteins, 60, 2005
|
|
6I4G
 
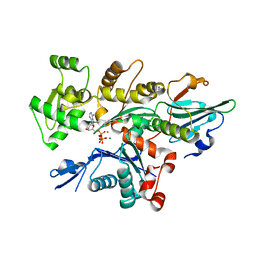 | | Crystal Structure of Plasmodium falciparum actin I (H74Q) in the Mg-K-ATP state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-1, BETA-MERCAPTOETHANOL, ... | | Authors: | Kumpula, E.-P, Lopez, A.J, Tajedin, L, Han, H, Kursula, I. | | Deposit date: | 2018-11-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic view into Plasmodium actin polymerization, ATP hydrolysis, and fragmentation.
Plos Biol., 17, 2019
|
|
6I6J
 
 | |
3A5L
 
 | | Crystal Structure of a Dictyostelium P109A Mg2+-Actin in Complex with Human Gelsolin Segment 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Gelsolin, ... | | Authors: | Murakami, K, Yasunaga, T, Noguchi, T.Q, Uyeda, T.Q, Wakabayashi, T. | | Deposit date: | 2009-08-09 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release
Cell(Cambridge,Mass.), 143, 2010
|
|
3AG1
 
 | | Bovine Heart Cytochrome c Oxidase in the Carbon Monoxide-bound Fully Reduced State at 280 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Ohta, K, Shinzawa-Itoh, K, Kanda, K, Taniguchi, M, Nabekura, H, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2010-03-19 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3AHR
 
 | | Inactive human Ero1 | | Descriptor: | ERO1-like protein alpha, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Inaba, K, Sitia, R, Suzuki, M. | | Deposit date: | 2010-04-26 | | Release date: | 2010-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Crystal structures of human Ero1-alpha reveal the mechanisms of regulated and targeted oxidation of PDI
Embo J., 29, 2010
|
|
1Y34
 
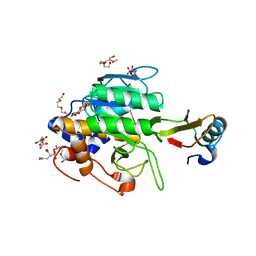 | | Crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 E60A mutant | | Descriptor: | CALCIUM ION, CITRIC ACID, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Radisky, E.S, Lu, C.J, Kwan, G, Koshland Jr, D.E. | | Deposit date: | 2004-11-23 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Role of the intramolecular hydrogen bond network in the inhibitory power of chymotrypsin inhibitor 2
Biochemistry, 44, 2005
|
|
2RM0
 
 | | FBP28WW2 domain in complex with a PPPLIPPPP peptide | | Descriptor: | Formin-1, Transcription elongation regulator 1 | | Authors: | Ramirez-Espain, X, Ruiz, L, Martin-Malpartida, P, Oschkinat, H, Macias, M.J. | | Deposit date: | 2007-09-06 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of a New Binding Motif and a Novel Binding Mode in Group 2 WW Domains
J.Mol.Biol., 373, 2007
|
|
3ML1
 
 | | Crystal Structure of the Periplasmic Nitrate Reductase from Cupriavidus necator | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DIOXOTHIOMOLYBDENUM(VI) ION, Diheme cytochrome c napB, ... | | Authors: | Coelho, C, Trincao, J, Romao, M.J. | | Deposit date: | 2010-04-16 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Cupriavidus necator nitrate reductase in oxidized and partially reduced states
J.Mol.Biol., 408, 2011
|
|
6I4M
 
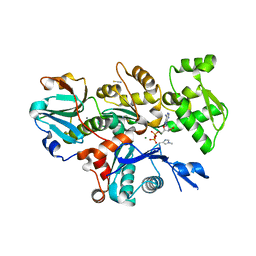 | | Crystal Structure of Plasmodium berghei actin II in the Mg-ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-2, CALCIUM ION, ... | | Authors: | Kumpula, E.-P, Lopez, A.J, Tajedin, L, Han, H, Kursula, I. | | Deposit date: | 2018-11-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.873 Å) | | Cite: | Atomic view into Plasmodium actin polymerization, ATP hydrolysis, and fragmentation.
Plos Biol., 17, 2019
|
|
6IB8
 
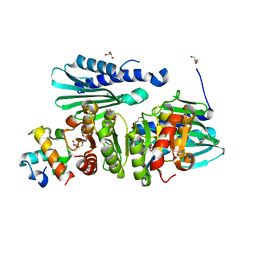 | | Structure of a complex of SuhB and NusA AR2 domain | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, Inositol-1-monophosphatase, ... | | Authors: | Huang, Y.H, Loll, B, Wahl, M.C. | | Deposit date: | 2018-11-29 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.646 Å) | | Cite: | Structural basis for the function of SuhB as a transcription factor in ribosomal RNA synthesis.
Nucleic Acids Res., 47, 2019
|
|
2RLY
 
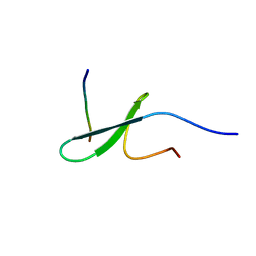 | | FBP28WW2 domain in complex with PTPPPLPP peptide | | Descriptor: | Formin-1, Transcription elongation regulator 1 | | Authors: | Ramirez-Espain, X, Ruiz, L, Martin-Malpartida, P, Oschkinat, H, Macias, M.J. | | Deposit date: | 2007-09-03 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of a New Binding Motif and a Novel Binding Mode in Group 2 WW Domains
J.Mol.Biol., 373, 2007
|
|
6I6X
 
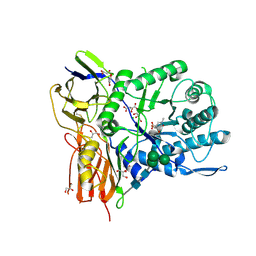 | | New Irreversible a-l-Iduronidase Inhibitors and Activity-Based Probes | | Descriptor: | (1~{R},2~{R},3~{R},4~{S},5~{S},6~{R})-7-methyl-3,4,5-tris(oxidanyl)-7-azabicyclo[4.1.0]heptane-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gloster, T.M, McMahon, S.A, Oehler, V. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | New Irreversible alpha-l-Iduronidase Inhibitors and Activity-Based Probes.
Chemistry, 24, 2018
|
|
6I9K
 
 | | Crystal structure of Jumping Spider Rhodopsin-1 bound to 9-cis retinal | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Kumopsin1, RETINAL | | Authors: | Varma, N, Mutt, E, Muehle, J, Panneels, V, Terakita, A, Deupi, X, Nogly, P, Schertler, F.X.G, Lesca, E. | | Deposit date: | 2018-11-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.145 Å) | | Cite: | Crystal structure of jumping spider rhodopsin-1 as a light sensitive GPCR.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8QBT
 
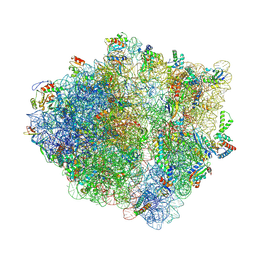 | | E. coli ApdP-stalled ribosomal complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S11, ... | | Authors: | Morici, M, Wilson, D.N. | | Deposit date: | 2023-08-25 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | RAPP-containing arrest peptides induce translational stalling by short circuiting the ribosomal peptidyltransferase activity.
Nat Commun, 15, 2024
|
|
6INF
 
 | | a glycosyltransferase complex with UDP | | Descriptor: | UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhu, X, Yang, T, Naismith, J.H. | | Deposit date: | 2018-10-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
1Y7A
 
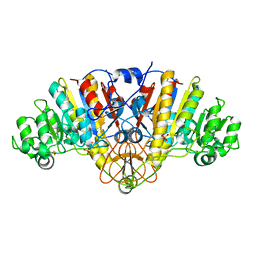 | | Structure of D153H/K328W E. coli alkaline phosphatase in presence of cobalt at 1.77 A resolution | | Descriptor: | Alkaline phosphatase, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Wang, J, Stieglitz, K, Kantrowitz, E.R. | | Deposit date: | 2004-12-08 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Metal Specificity Is Correlated with Two Crucial Active Site Residues in Escherichia coli Alkaline Phosphatase(,).
Biochemistry, 44, 2005
|
|
2XO5
 
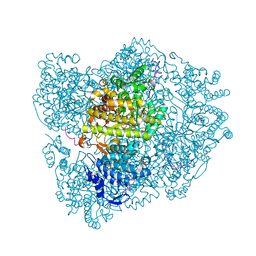 | | RIBONUCLEOTIDE REDUCTASE Y731NH2Y MODIFIED R1 SUBUNIT OF E. COLI | | Descriptor: | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT ALPHA, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT BETA | | Authors: | Minnihan, E.C, Seyedsayamdost, M.R, Uhlin, U, Stubbe, J. | | Deposit date: | 2010-08-09 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Kinetics of Radical Intermediate Formation and Deoxynucleotide Production in 3-Aminotyrosine- Substituted Escherichia Coli Ribonucleotide Reductases.
J.Am.Chem.Soc., 133, 2011
|
|
6IQL
 
 | | Crystal structure of dopamine receptor D4 bound to the subtype-selective ligand, L745870 | | Descriptor: | 3-{[4-(4-chlorophenyl)piperazin-1-yl]methyl}-1H-pyrrolo[2,3-b]pyridine, D(4) dopamine receptor,Soluble cytochrome b562,D(4) dopamine receptor | | Authors: | Zhou, Y, Cao, C, Zhang, X.C. | | Deposit date: | 2018-11-08 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of dopamine receptor D4 bound to the subtype selective ligand, L745870.
Elife, 8, 2019
|
|
3M29
 
 | |
2XP8
 
 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 4-(MORPHOLIN-4-YLCARBONYL)-2-PHENYL-1H-IMIDAZOLE-5-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3M3O
 
 | | Crystal Structure of Agrocybe aegerita lectin AAL mutant R85A complexed with p-Nitrophenyl TF disaccharide | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Anti-tumor lectin, P-NITROPHENOL, ... | | Authors: | Feng, L, Li, D, Wang, D. | | Deposit date: | 2010-03-09 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the recognition mechanism between an antitumor galectin AAL and the Thomsen-Friedenreich antigen
Faseb J., 24, 2010
|
|
