2NQC
 
 | | Crystal structure of ig-like domain 23 from human filamin C | | Descriptor: | Filamin-C, GLYCEROL, IMIDAZOLE, ... | | Authors: | Sjekloca, L, Pudas, R, Sjoeblom, B, Konarev, P, Carugo, O, Rybin, V, Kiema, T.R, Svergun, D, Ylanne, J, Djinovic-Carugo, K. | | Deposit date: | 2006-10-31 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of human filamin C domain 23 and small angle scattering model for filamin C 23-24 dimer
J.Mol.Biol., 368, 2007
|
|
3DN3
 
 | | Iodopentafluorobenzene binding in the hydrophobic cavity of T4 lysozyme L99A mutant | | Descriptor: | 1,2,3,4,5-pentafluoro-6-iodobenzene, 2-HYDROXYETHYL DISULFIDE, Lysozyme, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2008-07-01 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Halogenated benzenes bound within a non-polar cavity in T4 lysozyme provide examples of I...S and I...Se halogen-bonding.
J.Mol.Biol., 385, 2009
|
|
2NQI
 
 | | Calpain 1 proteolytic core inactivated by WR13(R,R), an epoxysuccinyl-type inhibitor. | | Descriptor: | CALCIUM ION, Calpain-1 catalytic subunit, N~2~-[(2S)-2-{[(2R)-4-ETHOXY-2-HYDROXY-4-OXOBUTANOYL]AMINO}PENT-4-ENOYL]-L-ARGINYL-L-TRYPTOPHANAMIDE | | Authors: | Cuerrier, D, Davies, P.L, Campbell, R.L, Moldoveanu, T. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Development of Calpain-specific Inactivators by Screening of Positional Scanning Epoxide Libraries
J.Biol.Chem., 282, 2007
|
|
3DQI
 
 | |
3DQU
 
 | |
5MRN
 
 | | Arabidopsis thaliana IspD Glu258Ala Mutant | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, CADMIUM ION, ... | | Authors: | Schwab, A, Illarionov, B, Frank, A, Kunfermann, A, Seet, M, Bacher, A, Witschel, M, Fischer, M, Groll, M, Diederich, F. | | Deposit date: | 2016-12-23 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of Allosteric Inhibition of the Enzyme IspD by Three Different Classes of Ligands.
ACS Chem. Biol., 12, 2017
|
|
8QC8
 
 | |
3DQJ
 
 | |
3DRU
 
 | | Crystal Structure of Gly117Phe Alpha1-Antitrypsin | | Descriptor: | Alpha-1-antitrypsin | | Authors: | Gooptu, B, Nobeli, I, Purkiss, A, Phillips, R.L, Mallya, M, Lomas, D.A, Barrett, T.E. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystallographic and cellular characterisation of two mechanisms stabilising the native fold of alpha1-antitrypsin: implications for disease and drug design.
J.Mol.Biol., 387, 2009
|
|
8QC2
 
 | | Crystal structure of NAD-dependent glycoside hydrolase from Flavobacterium sp. (strain K172) in complex with co-factor NAD+ and sulfoquinovose (SQ) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Gfo/Idh/MocA family oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Pickles, I.B, Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Widespread Family of NAD + -Dependent Sulfoquinovosidases at the Gateway to Sulfoquinovose Catabolism.
J.Am.Chem.Soc., 145, 2023
|
|
6SEF
 
 | | Class2C : CENP-A nucleosome in complex with CENP-C central region | | Descriptor: | Centromere protein C, DNA (145-MER), Histone H2A type 2-A, ... | | Authors: | Ali-Ahmad, A, Bilokapic, S, Schafer, I.B, Halic, M, Sekulic, N. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail.
Embo Rep., 20, 2019
|
|
1RP4
 
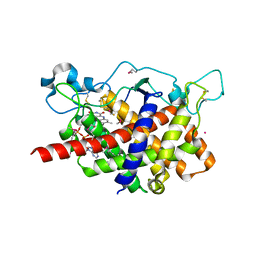 | | Structure of Ero1p, Source of Disulfide Bonds for Oxidative Protein Folding in the Cell | | Descriptor: | 1-ETHYL-PYRROLIDINE-2,5-DIONE, CADMIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Gross, E, Kastner, D.B, Kaiser, C.A, Fass, D. | | Deposit date: | 2003-12-03 | | Release date: | 2004-06-08 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of ero1p, source of disulfide bonds for oxidative protein folding in the cell.
Cell(Cambridge,Mass.), 117, 2004
|
|
7LVC
 
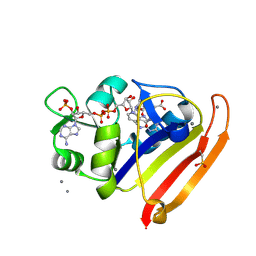 | | E. coli DHFR by Native Mn,P,S-SAD at Room Temperature | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Greisman, J.B, Dalton, K.M, Hekstra, D.R. | | Deposit date: | 2021-02-24 | | Release date: | 2021-03-17 | | Last modified: | 2022-08-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Native SAD phasing at room temperature.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1R4X
 
 | | Crystal Structure Analys of the Gamma-COPI Appendage domain | | Descriptor: | Coatomer gamma subunit, MAGNESIUM ION | | Authors: | Watson, P.J, Frigerio, G, Collins, B.M, Duden, R, Owen, D.J. | | Deposit date: | 2003-10-09 | | Release date: | 2003-10-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gamma-COP appendage domain - structure and function
Traffic, 5, 2004
|
|
4LNS
 
 | |
2NAO
 
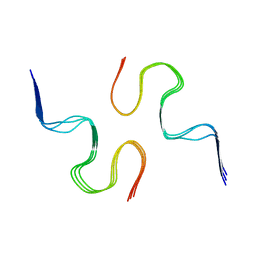 | | Atomic resolution structure of a disease-relevant Abeta(1-42) amyloid fibril | | Descriptor: | Beta-amyloid protein 42 | | Authors: | Waelti, M.A, Ravotti, F, Arai, H, Glabe, C, Wall, J, Bockmann, A, Guntert, P, Meier, B.H, Riek, R. | | Deposit date: | 2016-01-07 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Atomic-resolution structure of a disease-relevant A beta (1-42) amyloid fibril.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1RFD
 
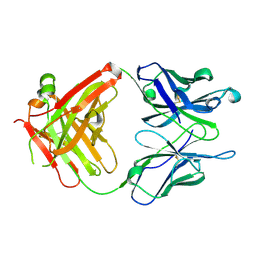 | | ANTI-COCAINE ANTIBODY M82G2 | | Descriptor: | Fab M82G2, Heavy Chain, Light Chain | | Authors: | Pozharski, E, Hewagama, A, Shanafelt, A.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2003-11-07 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Diversity in hapten recognition: structural study of an anti-cocaine antibody M82G2.
J.Mol.Biol., 349, 2005
|
|
3DJ7
 
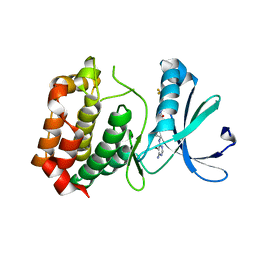 | | Crystal structure of the mouse Aurora-A catalytic domain (Asn186->Gly, Lys240->Arg, Met302->Leu) in complex with Compound 130. | | Descriptor: | 1-(5-{2-[(6-amino-5-bromopyrimidin-4-yl)amino]ethyl}-1,3-thiazol-2-yl)-3-[3-(trifluoromethyl)phenyl]urea, serine/threonine kinase 6 | | Authors: | Elling, R.A, Yang, W, Erlanson, D.A, Tangonan, B.T, Hansen, S.K, Romanowski, M.J. | | Deposit date: | 2008-06-22 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | New fragment-based drug discovery
To be Published
|
|
8QYF
 
 | |
3E1S
 
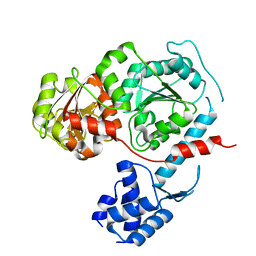 | | Structure of an N-terminal truncation of Deinococcus radiodurans RecD2 | | Descriptor: | Exodeoxyribonuclease V, subunit RecD | | Authors: | Saikrishnan, K, Griffiths, S.P, Cook, N, Court, R, Wigley, D.B. | | Deposit date: | 2008-08-04 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | DNA binding to RecD: role of the 1B domain in SF1B helicase activity.
Embo J., 27, 2008
|
|
5MG2
 
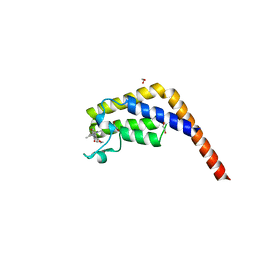 | | Crystal structure of the second bromodomain of human TAF1 in complex with BAY-299 chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 6-(3-oxidanylpropyl)-2-(1,3,6-trimethyl-2-oxidanylidene-benzimidazol-5-yl)benzo[de]isoquinoline-1,3-dione, Transcription initiation factor TFIID subunit 1 | | Authors: | Tallant, C, Bouche, L, Holton, S.J, Fedorov, O, Siejka, P, Picaud, S, Krojer, T, Srikannathasan, V, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hartung, I.V, Haendler, B, Muller, S, Huber, K.V.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Benzoisoquinolinediones as Potent and Selective Inhibitors of BRPF2 and TAF1/TAF1L Bromodomains.
J. Med. Chem., 60, 2017
|
|
5MD5
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=11, twist=0 | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MDD
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=23, twist=6 | | Descriptor: | Gag protein | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MIS
 
 | | Crystal Structure of Lactococcus lactis Thioredoxin Reductase Exposed to Visible Light (180 min) | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Skjoldager, N, Bang, M.B, Svensson, B, Hagglund, P, Harris, P. | | Deposit date: | 2016-11-29 | | Release date: | 2017-04-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The structure of Lactococcus lactis thioredoxin reductase reveals molecular features of photo-oxidative damage.
Sci Rep, 7, 2017
|
|
5MJ1
 
 | | Extracellular domain of human CD83 - rhombohedral crystal form | | Descriptor: | CD83 antigen, DI(HYDROXYETHYL)ETHER | | Authors: | Klingl, S, Egerer-Sieber, C, Schmid, B, Weiler, S, Muller, Y.A. | | Deposit date: | 2016-11-29 | | Release date: | 2017-03-29 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Extracellular Domain of the Human Dendritic Cell Surface Marker CD83.
J. Mol. Biol., 429, 2017
|
|
