5W2Q
 
 | | Crystal structure of Mycobacterium tuberculosis KasA in complex with 6U5 | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 1, GLYCEROL, ... | | Authors: | Capodagli, G.C, Neiditch, M.B. | | Deposit date: | 2017-06-06 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synergistic Lethality of a Binary Inhibitor of Mycobacterium tuberculosis KasA.
MBio, 9, 2018
|
|
3UH1
 
 | | Crystal Structure of Saccharopine Dehydrogenase from Saccharomyces cerevisiae with bound saccharopine and NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, N-(5-AMINO-5-CARBOXYPENTYL)GLUTAMIC ACID, ... | | Authors: | Kumar, V.P, Thomas, L.M, Bobyk, K.D, Andi, B, West, A.H, Cook, P.F. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Evidence in Support of Lysine 77 and Histidine 96 as Acid-Base Catalytic Residues in Saccharopine Dehydrogenase from Saccharomyces cerevisiae.
Biochemistry, 51, 2012
|
|
3UGK
 
 | | Crystal Structure of C205S mutant and Saccharopine Dehydrogenase from Saccharomyces cerevisiae. | | Descriptor: | Saccharopine dehydrogenase [NAD+, L-lysine-forming] | | Authors: | Cook, P.F, Kumar, V.P, Thomas, L.M, West, A.H, Bobyk, K.D. | | Deposit date: | 2011-11-02 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Evidence in Support of Lysine 77 and Histidine 96 as Acid-Base Catalytic Residues in Saccharopine Dehydrogenase from Saccharomyces cerevisiae.
Biochemistry, 51, 2012
|
|
7Z7T
 
 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z83
 
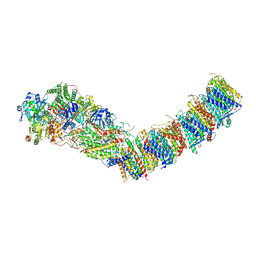 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z84
 
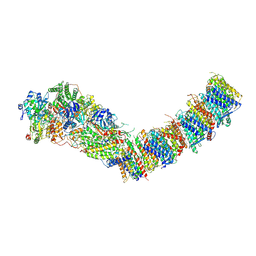 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Open-ready state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z7S
 
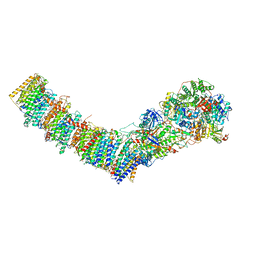 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Closed state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7ZC5
 
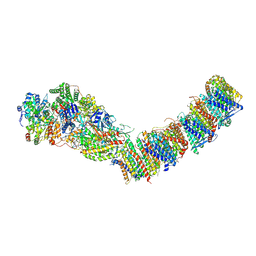 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-25 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
4P1W
 
 | |
1PO5
 
 | | Structure of mammalian cytochrome P450 2B4 | | Descriptor: | Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Scott, E.E, He, Y.A, Wester, M.R, White, M.A, Chin, C.C, Halpert, J.R, Johnson, E.F, Stout, C.D. | | Deposit date: | 2003-06-13 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An open conformation of mammalian cytochrome P450 2B4 at 1.6 A resolution
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
7Z80
 
 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Closed state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z7V
 
 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Open-ready state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
1PQ2
 
 | | Crystal Structure of Human Drug Metabolizing Cytochrome P450 2C8 | | Descriptor: | Cytochrome P450 2C8, PALMITIC ACID, PHOSPHATE ION, ... | | Authors: | Schoch, G.A, Yano, J.K, Wester, M.R, Griffin, K.J, Stout, C.D, Johnson, E.F. | | Deposit date: | 2003-06-17 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of human microsomal cytochrome P450 2C8. Evidence for a peripheral fatty acid binding site
J.Biol.Chem., 279, 2004
|
|
5ZXL
 
 | | Structure of GldA from E.coli | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycerol dehydrogenase, ... | | Authors: | Zhang, J, Lin, L. | | Deposit date: | 2018-05-21 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Structure of glycerol dehydrogenase (GldA) from Escherichia coli.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
7Z7R
 
 | | Complex I from E. coli, LMNG-purified, Apo, Open-ready state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7ZCI
 
 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-28 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z0T
 
 | | Structure of the Escherichia coli formate hydrogenlyase complex (aerobic preparation, composite structure) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CARBONMONOXIDE-(DICYANO) IRON, FE (III) ION, ... | | Authors: | Steinhilper, R, Murphy, B.J. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-28 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the membrane-bound formate hydrogenlyase complex from Escherichia coli.
Nat Commun, 13, 2022
|
|
6AHC
 
 | |
2YYL
 
 | | Crystal structure of the mutant of HpaB (T198I, A276G, and R466H) complexed with FAD | | Descriptor: | 4-hydroxyphenylacetate-3-hydroxylase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-04-30 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
3UHA
 
 | | Crystal Structure of Saccharopine Dehydrogenase from Saccharomyces cervisiae complexed with NAD. | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Saccharopine dehydrogenase [NAD+, ... | | Authors: | Cook, P.F, Kumar, V.P, Thomas, L.M, West, A.H, Bobyk, K.D. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence in Support of Lysine 77 and Histidine 96 as Acid-Base Catalytic Residues in Saccharopine Dehydrogenase from Saccharomyces cerevisiae.
Biochemistry, 51, 2012
|
|
4NMQ
 
 | |
4NMV
 
 | |
3UHF
 
 | | Crystal Structure of Glutamate Racemase from Campylobacter jejuni subsp. jejuni | | Descriptor: | CHLORIDE ION, D-GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Maltseva, N, Mulligan, R, Kwon, K, Kim, Y, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-03 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Glutamate Racemase
from Campylobacter jejuni subsp. jejuni
To be Published
|
|
3S81
 
 | | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium | | Descriptor: | CHLORIDE ION, Putative aspartate racemase, SULFATE ION | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Zhang, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-27 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium
To be Published
|
|
4NF9
 
 | | Structure of the Knl1/Nsl1 complex | | Descriptor: | CHLORIDE ION, Kinetochore-associated protein NSL1 homolog, Protein CASC5 | | Authors: | Petrovic, A, Mosalaganti, S, Keller, J, Mattiuzzo, M, Overlack, K, Wohlgemuth, S, Pasqualato, S, Raunser, S, Musacchio, A. | | Deposit date: | 2013-10-31 | | Release date: | 2014-03-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Modular Assembly of RWD Domains on the Mis12 Complex Underlies Outer Kinetochore Organization.
Mol.Cell, 53, 2014
|
|
