8IT4
 
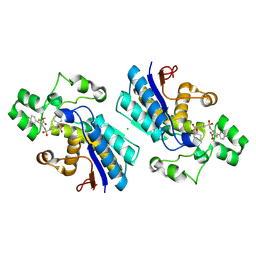 | | Phosphoglycerate mutase 1 complexed with a covalent inhibitor | | Descriptor: | 3-[[5-(4-chlorophenyl)-2-methoxycarbonyl-thiophen-3-yl]sulfamoyl]benzenesulfonic acid, CHLORIDE ION, Phosphoglycerate mutase 1 | | Authors: | Zhou, L, Jiang, L.L. | | Deposit date: | 2023-03-22 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phosphoglycerate mutase 1 complexed with a covalent inhibitor
To Be Published
|
|
8IT6
 
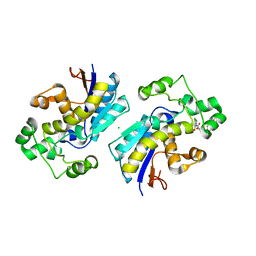 | | Phosphoglycerate mutase 1 complexed with a compound | | Descriptor: | 4-[4-chloranyl-2-(2-hydroxy-2-oxoethyloxy)phenyl]-2-[(4-phenylphenyl)sulfonylamino]benzoic acid, CHLORIDE ION, Phosphoglycerate mutase 1 | | Authors: | Zhou, L, Jiang, L.L. | | Deposit date: | 2023-03-22 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Phosphoglycerate mutase 1 complexed with a compound
To Be Published
|
|
8ITB
 
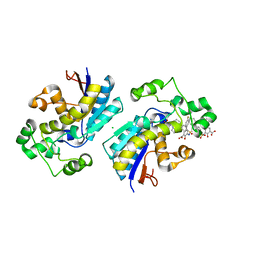 | | Phosphoglycerate mutase 1 complexed with a compound | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(3-chloranyl-4-fluoranyl-phenyl)-2-oxidanyl-3-[(4-phenylphenyl)sulfonylamino]benzoic acid, CHLORIDE ION, ... | | Authors: | Zhou, L, Jiang, L.L. | | Deposit date: | 2023-03-22 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Phosphoglycerate mutase 1 complexed with a compound
To Be Published
|
|
8IT5
 
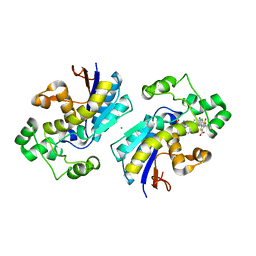 | | Phosphoglycerate mutase 1 complexed with a compound | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(4-chlorophenyl)-3-[(4-phenylphenyl)sulfonylamino]thiophene-2-carboxylic acid, CHLORIDE ION, ... | | Authors: | Zhou, L, Jiang, L.L. | | Deposit date: | 2023-03-22 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phosphoglycerate mutase 1 complexed with a compound
To Be Published
|
|
8TX8
 
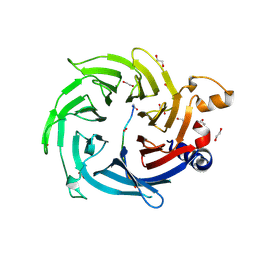 | | Crystal Structure of RBBP4 bound to ZNF512B peptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FORMIC ACID, ... | | Authors: | Deshpande, C.N, Mackay, J.P. | | Deposit date: | 2023-08-22 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ZNF512B binds RBBP4 via a variant NuRD interaction motif and aggregates chromatin in a NuRD complex-independent manner.
Nucleic Acids Res., 52, 2024
|
|
8XPE
 
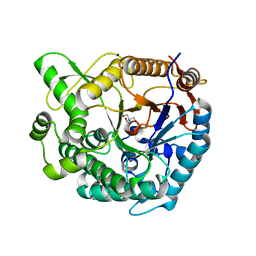 | | Crystal structure of Tris-bound TsaBgl (DATA III) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, beta-glucosidase | | Authors: | Nam, K.H. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of Tris binding in beta-glucosidases.
Biochem.Biophys.Res.Commun., 700, 2024
|
|
8XPC
 
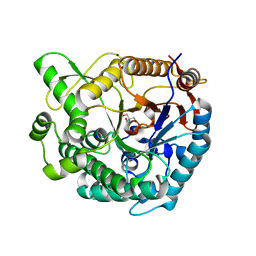 | | Crystal structure of Tris-bound TsaBgl (DATA I) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, beta-glucosidase | | Authors: | Nam, K.H. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of Tris binding in beta-glucosidases.
Biochem.Biophys.Res.Commun., 700, 2024
|
|
8XPD
 
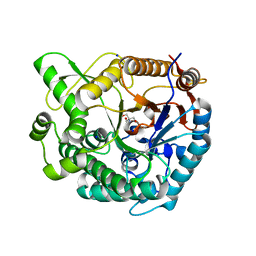 | | Crystal structure of Tris-bound TsaBgl (DATA II) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, beta-glucosidase | | Authors: | Nam, K.H. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of Tris binding in beta-glucosidases.
Biochem.Biophys.Res.Commun., 700, 2024
|
|
7U4M
 
 | | Crystal structure of human GPX4-U46C in complex with LOC1886 | | Descriptor: | 1,2-ETHANEDIOL, 4-methoxy-1H-indole-2-carbaldehyde, Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
5Y5I
 
 | | Time-resolved SFX structure of cytochrome P450nor: 20 ms after photo-irradiation of caged NO in the presence of NADH (NO-bound state), light data | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
6IWK
 
 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 | | Descriptor: | CALCIUM ION, GLYCEROL, Glucan 1,4-alpha-maltotetraohydrolase | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structure of maltotetraose-forming amylase from Pseudomonas saccharophila STB07 provides insights into its product specificity.
Int.J.Biol.Macromol., 154, 2020
|
|
8WEY
 
 | | PSI-LHCI of the red alga Cyanidium caldarium RK-1 (NIES-2137) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Kato, K, Hamaguchi, T, Nakajima, Y, Kawakami, K, Yonekura, K, Shen, J.R, Nagao, R. | | Deposit date: | 2023-09-19 | | Release date: | 2024-02-14 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (1.92 Å) | | Cite: | The structure of PSI-LHCI from Cyanidium caldarium provides evolutionary insights into conservation and diversity of red-lineage LHCs.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7AOB
 
 | | Crystal structure of Thermaerobacter marianensis malate dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Bertrand, Q, Lasalle, L, Girard, E, Madern, D. | | Deposit date: | 2020-10-14 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of Thermaerobacter marianensis malate dehydrogenase
To Be Published
|
|
6XLS
 
 | |
9MG0
 
 | | Structure of Kluyveromyces lactis mRNA cap (guanine-N7) methyltransferase, Abd1, in complex with SAH | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nilson, D.J, Fedorov, E, Ghosh, A. | | Deposit date: | 2024-12-10 | | Release date: | 2025-07-09 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for sensitivity and acquired resistance of fungal cap guanine-N7 methyltransferases to the antifungal antibiotic sinefungin.
Nucleic Acids Res., 53, 2025
|
|
6RQZ
 
 | | GOLGI ALPHA-MANNOSIDASE II complex with Manno-epi-cyclophellitol aziridine | | Descriptor: | (1~{R},2~{S},3~{R},4~{R},5~{S},6~{R})-5-azanyl-6-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Alpha-mannosidase 2, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, Debets, M, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-16 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
5Y5J
 
 | | Time-resolved SFX structure of cytochrome P450nor: dark-2 data in the presence of NADH (resting state) | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
6GTA
 
 | | Alpha-galactosidase mutant D378A from Thermotoga maritima in complex with intact cyclohexene-based carbasugar mimic of galactose with 3,5 difluorophenyl leaving group | | Descriptor: | (1~{R},2~{S},3~{S},6~{S})-6-[3,5-bis(fluoranyl)phenoxy]-4-(hydroxymethyl)cyclohex-4-ene-1,2,3-triol, Alpha-galactosidase, MAGNESIUM ION, ... | | Authors: | Gloster, T.M, Pengelly, R.J. | | Deposit date: | 2018-06-17 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Revealing the mechanism for covalent inhibition of glycoside hydrolases by carbasugars at an atomic level.
Nat Commun, 9, 2018
|
|
7VBE
 
 | |
5O1O
 
 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain with proline bound. | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, mitochondrial, ... | | Authors: | Kopec, J, Rembeza, E, Pena, I.A, Mathea, S, Velupillai, S, Strain-Damerell, C, Goubin, S, Kupinska, K, Talon, R, Collins, P, Krojer, T, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, von Delft, F, Arruda, P, Yue, W.W. | | Deposit date: | 2017-05-18 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain with proline bound.
To Be Published
|
|
9MG4
 
 | | Structure of Saccharomyces cerevisiae mRNA cap (guanine-N7) methyltransferase, Abd1, in complex with SAH | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Nilson, D.J, Fedorov, E, Ghosh, A. | | Deposit date: | 2024-12-10 | | Release date: | 2025-07-09 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for sensitivity and acquired resistance of fungal cap guanine-N7 methyltransferases to the antifungal antibiotic sinefungin.
Nucleic Acids Res., 53, 2025
|
|
6H1T
 
 | | Structure of the BM3 heme domain in complex with clotrimazole | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, ... | | Authors: | Jeffreys, L.N, Munro, A.W.M, Leys, D. | | Deposit date: | 2018-07-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Novel insights into P450 BM3 interactions with FDA-approved antifungal azole drugs.
Sci Rep, 9, 2019
|
|
9DC9
 
 | | Mtb GuaB dCBS in complex with inhibitor compound 5 | | Descriptor: | (3P)-3-(4-chloro-1H-imidazol-2-yl)-5-[(1R)-1-{[(3M)-3-(1H-pyrazol-5-yl)phenyl]oxy}ethyl]pyridine, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-08-25 | | Release date: | 2024-11-27 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of potent dihydro-oxazinoquinolinone inhibitors of GuaB for the treatment of tuberculosis.
Bioorg.Med.Chem.Lett., 117, 2025
|
|
9DC8
 
 | | Mtb GuaB dCBS in complex with inhibitor G1 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-(6-chloropyridin-3-yl)-N~2~-(1,4-dihydro-2H-pyrano[3,4-c]quinolin-9-yl)-L-alaninamide, ... | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-08-25 | | Release date: | 2024-11-27 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of potent dihydro-oxazinoquinolinone inhibitors of GuaB for the treatment of tuberculosis.
Bioorg.Med.Chem.Lett., 117, 2025
|
|
8OWM
 
 | | Crystal structure of glutamate dehydrogenase 2 from Arabidopsis thaliana binding Ca, NAD and 2,2-dihydroxyglutarate | | Descriptor: | 1,2-ETHANEDIOL, 2,2-bis(oxidanyl)pentanedioic acid, CALCIUM ION, ... | | Authors: | Grzechowiak, M, Ruszkowski, M. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional studies of Arabidopsis thaliana glutamate dehydrogenase isoform 2 demonstrate enzyme dynamics and identify its calcium binding site.
Plant Physiol Biochem., 201, 2023
|
|
