3GR9
 
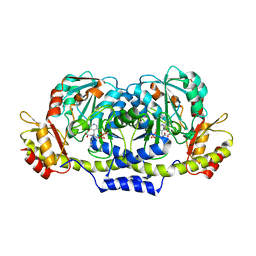 | | Crystal structure of ColD H188K S187N | | Descriptor: | 2-OXOGLUTARIC ACID, ColD | | Authors: | Holden, H.M, Cook, P.D, Kubiak, R.L, Toomey, D.P. | | Deposit date: | 2009-03-25 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two Site-Directed Mutations Are Required for the Conversion of a Sugar Dehydratase into an Aminotransferase.
Biochemistry, 48, 2009
|
|
3GST
 
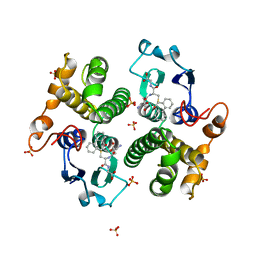 | | STRUCTURE OF THE XENOBIOTIC SUBSTRATE BINDING SITE OF A GLUTATHIONE S-TRANSFERASE AS REVEALED BY X-RAY CRYSTALLOGRAPHIC ANALYSIS OF PRODUCT COMPLEXES WITH THE DIASTEREOMERS OF 9-(S-GLUTATHIONYL)-10-HYDROXY-9, 10-DIHYDROPHENANTHRENE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, GLUTATHIONE S-TRANSFERASE, SULFATE ION | | Authors: | Ji, X, Ammon, H.L, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1993-06-07 | | Release date: | 1993-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the xenobiotic substrate binding site of a glutathione S-transferase as revealed by X-ray crystallographic analysis of product complexes with the diastereomers of 9-(S-glutathionyl)-10-hydroxy-9,10-dihydrophenanthrene.
Biochemistry, 33, 1994
|
|
4DAJ
 
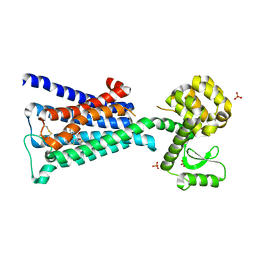 | | Structure of the M3 Muscarinic Acetylcholine Receptor | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, Muscarinic acetylcholine receptor M3, Lysozyme, ... | | Authors: | Kruse, A.C, Hu, J, Pan, A.C, Arlow, D.H, Rosenbaum, D.M, Rosemond, E, Green, H.F, Liu, T, Chae, P.S, Dror, R.O, Shaw, D.E, Weis, W.I, Wess, J, Kobilka, B. | | Deposit date: | 2012-01-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and dynamics of the M3 muscarinic acetylcholine receptor.
Nature, 482, 2012
|
|
4DBL
 
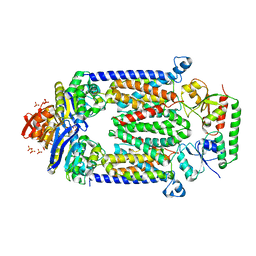 | | Crystal structure of E159Q mutant of BtuCDF | | Descriptor: | PHOSPHATE ION, SULFATE ION, Vitamin B12 import ATP-binding protein BtuD, ... | | Authors: | Korkhov, V.M, Mireku, S.M, Hvorup, R.N, Locher, K.P. | | Deposit date: | 2012-01-16 | | Release date: | 2012-03-07 | | Last modified: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (3.493 Å) | | Cite: | Asymmetric states of vitamin B12 transporter BtuCD are not discriminated by its cognate substrate binding protein BtuF.
Febs Lett., 586, 2012
|
|
2LTW
 
 | | YAP WW1 in complex with a Smad7 derived peptide | | Descriptor: | Smad7 derived peptide, Yorkie homolog | | Authors: | Macias, M.J, Aragon, E, Goerner, N, Xi, Q, Lopes, T, Gao, S, Massague, J. | | Deposit date: | 2012-06-04 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Versatile Interactions of Smad7 with Regulator WW Domains in TGF-beta Pathways.
Structure, 20, 2012
|
|
2L91
 
 | |
2L2B
 
 | |
2LAW
 
 | | Structure of the second WW domain from human YAP in complex with a human Smad1 derived peptide | | Descriptor: | Mothers against decapentaplegic homolog 1, Yorkie homolog | | Authors: | Macias, M.J, Aragon, E, Goerner, N, Zaromytidou, A, Xi, Q, Escobedo, A, Massague, J. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Smad action turnover switch operated by WW domain readers of a phosphoserine code.
Genes Dev., 25, 2011
|
|
2L5F
 
 | | Solution structure of the tandem WW domains from HYPA/FBP11 | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Jiang, Y, Hu, H. | | Deposit date: | 2010-11-01 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Interaction with polyglutamine expanded huntingtin alters cellular distribution and RNA processing of huntingtin yeast two-hybrid protein A (HYPA)
To be Published
|
|
2PMQ
 
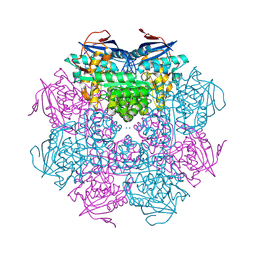 | | Crystal structure of a mandelate racemase/muconate lactonizing enzyme from Roseovarius sp. HTCC2601 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Lau, C, Sridhar, V, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of new enzymes and metabolic pathways by using structure and genome context.
Nature, 502, 2013
|
|
2PUJ
 
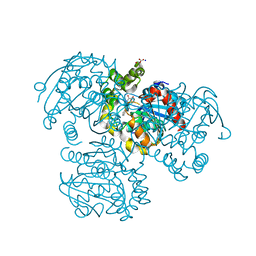 | | Crystal Structure of the S112A/H265A double mutant of a C-C hydrolase, BphD from Burkholderia xenovorans LB400, in complex with its substrate HOPDA | | Descriptor: | (2E,4E)-2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOIC ACID, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, MALONATE ION, ... | | Authors: | Bhowmik, S, Bolin, J.T. | | Deposit date: | 2007-05-09 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The Tautomeric Half-reaction of BphD, a C-C Bond Hydrolase: KINETIC AND STRUCTURAL EVIDENCE SUPPORTING A KEY ROLE FOR HISTIDINE 265 OF THE CATALYTIC TRIAD.
J.Biol.Chem., 282, 2007
|
|
2PZF
 
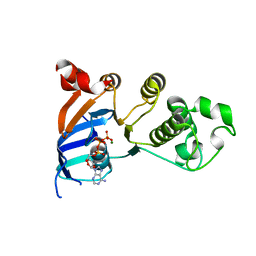 | | Minimal human CFTR first nucleotide binding domain as a head-to-tail dimer with delta F508 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Atwell, S, Conners, K, Emtage, S, Gheyi, T, Glenn, N.R, Hendle, J, Lewis, H.A, Lu, F, Rodgers, L.A, Romero, R, Sauder, J.M, Smith, D, Tien, H, Wasserman, S.R, Zhao, X. | | Deposit date: | 2007-05-18 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of a minimal human CFTR first nucleotide-binding domain as a monomer, head-to-tail homodimer, and pathogenic mutant.
Protein Eng.Des.Sel., 23, 2010
|
|
2N0G
 
 | | NMR structure of Neuromedin C in 90% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2MPR
 
 | | MALTOPORIN FROM SALMONELLA TYPHIMURIUM | | Descriptor: | CALCIUM ION, MALTOPORIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Meyer, J.E.W, Schulz, G.E. | | Deposit date: | 1997-02-07 | | Release date: | 1997-04-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of maltoporin from Salmonella typhimurium ligated with a nitrophenyl-maltotrioside.
J.Mol.Biol., 266, 1997
|
|
2MXV
 
 | |
2N1O
 
 | |
2N2L
 
 | | NMR structure of yersinia pestis ail (attachment invasion locus) in decylphosphocholine micelles calculated with implicit membrane solvation | | Descriptor: | Outer membrane protein X | | Authors: | Marassi, F.M, Ding, Y, Tian, Y, Schwieters, C.D, Yao, Y. | | Deposit date: | 2015-05-10 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Backbone structure of Yersinia pestis Ail determined in micelles by NMR-restrained simulated annealing with implicit membrane solvation.
J.Biomol.Nmr, 63, 2015
|
|
2MWB
 
 | | FBP28 WW2 mutant W457F | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Macias, M.J, Scheraga, H, Sunol, D, Todorovski, T. | | Deposit date: | 2014-11-03 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding kinetics of WW domains with the united residue force field for bridging microscopic motions and experimental measurements.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2NC6
 
 | | Solution Structure of N-L-idosylated Pin1 WW Domain | | Descriptor: | Pin1 WW Domain, beta-L-idopyranose | | Authors: | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | Deposit date: | 2016-03-20 | | Release date: | 2016-06-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
2MWD
 
 | |
2N4U
 
 | |
2N4R
 
 | |
2N1P
 
 | |
2MUQ
 
 | | Solution Structure of the Human FAAP20 UBZ | | Descriptor: | Fanconi anemia-associated protein of 20 kDa, ZINC ION | | Authors: | Wojtaszek, J.L, Wang, S, Zhou, P. | | Deposit date: | 2014-09-16 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin recognition by FAAP20 expands the complex interface beyond the canonical UBZ domain.
Nucleic Acids Res., 42, 2014
|
|
2N2M
 
 | |
