1W6J
 
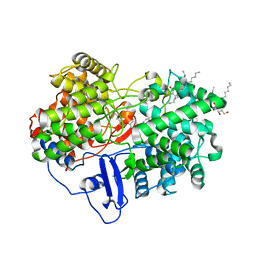 | | Structure of human OSC in complex with Ro 48-8071 | | Descriptor: | LANOSTEROL SYNTHASE, TETRADECANE, [4-({6-[ALLYL(METHYL)AMINO]HEXYL}OXY)-2-FLUOROPHENYL](4-BROMOPHENYL)METHANONE, ... | | Authors: | Thoma, R, Schulz-Gasch, T, D'Arcy, B, Benz, J, Aebi, J, Dehmlow, H, Hennig, M, Ruf, A. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insight Into Steroid Scaffold Formation from the Structure of Human Oxidosqualene Cyclase
Nature, 432, 2004
|
|
7WSB
 
 | | The ternary complex structure of FtmOx1 with a-ketoglutarate and 13-oxo-fumitremorgin B | | Descriptor: | 13-Oxofumitremorgin B, 2-OXOGLUTARIC ACID, COBALT (II) ION, ... | | Authors: | Wang, J, Wang, X.Y, Wang, Y.Y, Yan, W.P. | | Deposit date: | 2022-01-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Dissecting the Mechanism of the Nonheme Iron Endoperoxidase FtmOx1 Using Substrate Analogues.
Jacs Au, 2, 2022
|
|
7ZOE
 
 | |
6QLG
 
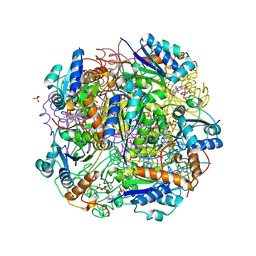 | | Crystal structure of AnUbiX (PadA1) in complex with FMN and dimethylallyl pyrophosphate | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYLALLYL DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Marshall, S.A, Payne, K.A.P, Leys, D. | | Deposit date: | 2019-02-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The UbiX flavin prenyltransferase reaction mechanism resembles class I terpene cyclase chemistry.
Nat Commun, 10, 2019
|
|
8A4G
 
 | |
6GSP
 
 | | RIBONUCLEASE T1/3'-GMP, 15 WEEKS | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Zegers, I, Wyns, L. | | Deposit date: | 1997-12-09 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hydrolysis of a slow cyclic thiophosphate substrate of RNase T1 analyzed by time-resolved crystallography.
Nat.Struct.Biol., 5, 1998
|
|
6O9N
 
 | | Structural insights on a new fungal aryl-alcohol oxidase | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kadowaki, M.A.S, Polikarpov, I. | | Deposit date: | 2019-03-14 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Enzymatic versatility and thermostability of a new aryl-alcohol oxidase from Thermothelomyces thermophilus M77.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
7VZS
 
 | | FAD-dpendent Glucose Dehydrogenase complexed with an inhibitor at pH7.56 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-glucal, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Nakajima, Y. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational change of catalytic residue in reduced enzyme of FAD-dependent Glucose Dehydrogenase at pH6.5
To Be Published
|
|
3IIA
 
 | | Crystal structure of apo (91-244) RIa subunit of cAMP-dependent protein kinase | | Descriptor: | GLYCEROL, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Sjoberg, T.J, Kim, C, Kornev, A.P, Taylor, S.S. | | Deposit date: | 2009-07-31 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cyclic AMP analog blocks kinase activation by stabilizing inactive conformation: conformational selection highlights a new concept in allosteric inhibitor design.
Mol.Cell Proteomics, 10, 2011
|
|
7WHE
 
 | |
7WKM
 
 | |
7WH6
 
 | | The mutant crystal structure of b-1,4-Xylanase (XynAF1_N179S) | | Descriptor: | Beta-xylanase, GLYCEROL, alpha-D-mannopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, G.Q, Zhang, R.F. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The mutant crystal structure of b-1,4-Xylanase (XynAF1_N179S)
To Be Published
|
|
7WPY
 
 | | AndA_M119A_N121V variant | | Descriptor: | Dioxygenase andA, FE (III) ION | | Authors: | Mori, T, Chen, H, Abe, I. | | Deposit date: | 2022-01-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | AndA_M119A_N121V variant
To Be Published
|
|
7XAY
 
 | | Crystal structure of Hat1-Hat2-Asf1-H3-H4 | | Descriptor: | COENZYME A, Histone H3, Histone H4, ... | | Authors: | Yue, Y, Yang, W.S, Xu, R.M. | | Deposit date: | 2022-03-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Topography of histone H3-H4 interaction with the Hat1-Hat2 acetyltransferase complex.
Genes Dev., 36, 2022
|
|
7WH7
 
 | | The mutant crystal structure of b-1,4-Xylanase (XynAF1_N179S) with xylotetraose | | Descriptor: | Beta-xylanase, alpha-D-mannopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, G.Q, Zhang, R.F. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | The mutant crystal structure of b-1,4-Xylanase (XynAF1_N179S) with xylotetraose
To Be Published
|
|
1OC6
 
 | | structure native of the D405N mutant of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS at 1.5 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CELLOBIOHYDROLASE II, ... | | Authors: | Varrot, A, Frandsen, T.P, Von Ossowski, I, Boyer, V, Driguez, H, Schulein, M, Davies, G.J. | | Deposit date: | 2003-02-06 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Ligand Binding and Processivity in Cellobiohydrolase Cel6A from Humicola Insolens
Structure, 11, 2003
|
|
7WHA
 
 | |
1NKO
 
 | | Energetic and structural basis of sialylated oligosaccharide recognition by the natural killer cell inhibitory receptor p75/AIRM1 or Siglec-7 | | Descriptor: | Sialic acid binding Ig-like lectin 7 | | Authors: | Dimasi, N, Attril, H, van Aalten, D.M.F, Moretta, L, Biassoni, R, Mariuzza, R.A. | | Deposit date: | 2003-01-03 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the saccharide-binding domain of the human natural killer cell inhibitory receptor p75/AIRM1.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1VM0
 
 | | X-RAY STRUCTURE OF GENE PRODUCT FROM ARABIDOPSIS THALIANA AT2G34160 | | Descriptor: | NITRATE ION, POTASSIUM ION, unknown protein | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-08-24 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-RAY STRUCTURE OF GENE PRODUCT FROM ARABIDOPSIS THALIANA AT2G34160
To be published
|
|
1VKP
 
 | | X-RAY STRUCTURE OF GENE PRODUCT FROM ARABIDOPSIS THALIANA AT5G08170, AGMATINE IMINOHYDROLASE | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, AGMATINE IMINOHYDROLASE, ... | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-06-15 | | Release date: | 2004-08-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g08170
To be published
|
|
2F2G
 
 | | X-Ray Structure of Gene Product From Arabidopsis Thaliana AT3G16990 | | Descriptor: | 4-AMINO-5-HYDROXYMETHYL-2-METHYLPYRIMIDINE, SEED MATURATION PROTEIN PM36 HOMOLOG, SULFATE ION | | Authors: | Wesenberg, G.W, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-11-16 | | Release date: | 2005-12-13 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of gene locus At3g16990 from Arabidopsis thaliana
Proteins, 57, 2004
|
|
4BU4
 
 | | RIBONUCLEASE T1 COMPLEX WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Loris, R, Devos, S, Langhorst, U, Decanniere, K, Bouckaert, J, Maes, D, Transue, T.R, Steyaert, J. | | Deposit date: | 1998-09-14 | | Release date: | 1998-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conserved water molecules in a large family of microbial ribonucleases.
Proteins, 36, 1999
|
|
7NF3
 
 | | Structure of A. niger Fdc T395M variant (AnFdcI) in complex with prFMN | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Saaret, A, Leys, D. | | Deposit date: | 2021-02-05 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Directed evolution of prenylated FMN-dependent Fdc supports efficient in vivo isobutene production.
Nat Commun, 12, 2021
|
|
7NF4
 
 | |
7NIN
 
 | | X-ray crystal structure of LsAA9A - CinnamtanninB1 soak | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Auxiliary activity 9, ... | | Authors: | Frandsen, K.E.H, Tokin, R, Skov, L, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-02-12 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of lytic polysaccharide monooxygenase by natural plant extracts.
New Phytol., 232, 2021
|
|
