1O0H
 
 | | Ribonuclease A in complex with 5'-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ribonuclease pancreatic | | Authors: | Leonidas, D.D, Oikonomakos, N.G, Chrysina, E.D, Kosmopoulou, M.N, Vlassi, M. | | Deposit date: | 2003-02-21 | | Release date: | 2003-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution crystal structures of ribonuclease A complexed with adenylic and uridylic nucleotide inhibitors. Implications for structure-based design of ribonucleolytic inhibitors
PROTEIN SCI., 12, 2003
|
|
1O0K
 
 | | Structure of the First Parallel DNA Quadruplex-drug Complex | | Descriptor: | 5'-D(*TP*GP*GP*GP*GP*T)-3', DAUNOMYCIN, SODIUM ION | | Authors: | Clark, G.R, Pytel, P.D, Squire, C.J, Neidle, S. | | Deposit date: | 2003-02-22 | | Release date: | 2003-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structure of the First Parallel DNA Quadruplex-drug Complex
J.Am.Chem.Soc., 125, 2003
|
|
1O0L
 
 | | THE STRUCTURE OF BCL-W REVEALS A ROLE FOR THE C-TERMINAL RESIDUES IN MODULATING BIOLOGICAL ACTIVITY | | Descriptor: | Apoptosis regulator Bcl-W | | Authors: | Hinds, M.G, Lackmann, M, Skea, G.L, Harrison, P.J, Huang, D.C.S, Day, C.L. | | Deposit date: | 2003-02-22 | | Release date: | 2003-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of Bcl-w reveals a role for the C-terminal residues in modulating biological activity
Embo J., 22, 2003
|
|
1O0M
 
 | | Ribonuclease A in complex with uridine-2'-phosphate | | Descriptor: | PHOSPHORIC ACID MONO-[2-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-4-HYDROXY-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-YL] ESTER, Ribonuclease pancreatic | | Authors: | Leonidas, D.D, Oikonomakos, N.G, Chrysina, E.D, Kosmopoulou, M.N, Vlassi, M. | | Deposit date: | 2003-02-24 | | Release date: | 2003-12-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structures of ribonuclease A complexed with adenylic and uridylic nucleotide inhibitors. Implications for structure-based design of ribonucleolytic inhibitors
PROTEIN SCI., 12, 2003
|
|
1O0N
 
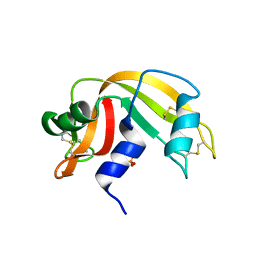 | | Ribonuclease A in complex with uridine-3'-phosphate | | Descriptor: | 3'-URIDINEMONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | Leonidas, D.D, Oikonomakos, N.G, Chrysina, E.D, Kosmopoulou, M.N, Vlassi, M. | | Deposit date: | 2003-02-24 | | Release date: | 2003-12-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structures of ribonuclease A complexed with adenylic and uridylic nucleotide inhibitors. Implications for structure-based design of ribonucleolytic inhibitors
PROTEIN SCI., 12, 2003
|
|
1O0O
 
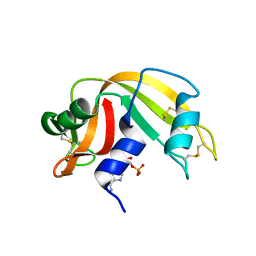 | | Ribonuclease A in complex with adenosine-2',5'-diphosphate | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, Ribonuclease pancreatic | | Authors: | Leonidas, D.D, Oikonomakos, N.G, Chrysina, E.D, Kosmopoulou, M.N, Vlassi, M. | | Deposit date: | 2003-02-24 | | Release date: | 2003-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution crystal structures of ribonuclease A complexed with adenylic and uridylic nucleotide inhibitors. Implications for structure-based design of ribonucleolytic inhibitors
PROTEIN SCI., 12, 2003
|
|
1O0P
 
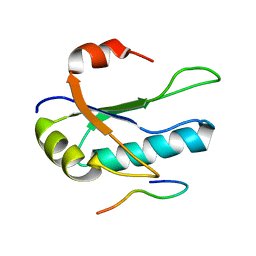 | | Solution Structure of the third RNA Recognition Motif (RRM) of U2AF65 in complex with an N-terminal SF1 peptide | | Descriptor: | Splicing Factor SF1, Splicing factor U2AF 65 kDa subunit | | Authors: | Selenko, P, Gregorovic, G, Sprangers, R, Stier, G, Rhani, Z, Kramer, A, Sattler, M. | | Deposit date: | 2003-02-24 | | Release date: | 2003-05-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP
Mol.Cell, 11, 2003
|
|
1O0Q
 
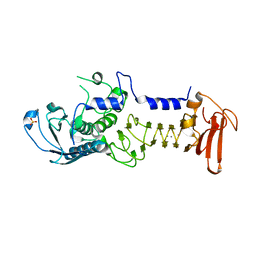 | | Crystal structure of a cold adapted alkaline protease from Pseudomonas TAC II 18, co-crystallized with 1 mM EDTA | | Descriptor: | CALCIUM ION, SULFATE ION, serralysin | | Authors: | Ravaud, S, Gouet, P, Haser, R, Aghajari, N. | | Deposit date: | 2003-02-24 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the role of divalent metal ions in a bacterial psychrophilic metalloprotease: binding studies of an enzyme in the crystalline state by x-ray crystallography.
J.Bacteriol., 185, 2003
|
|
1O0R
 
 | | Crystal structure of the catalytic domain of bovine beta1,4-galactosyltransferase complex with UDP-galactose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GALACTOSE-URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Ramakrishnan, B, Balaji, P.V, Qasba, P.K. | | Deposit date: | 2003-02-24 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF BETA 1,4-GALACTOSYLTRANSFERASE COMPLEX WITH UDP-GAL
REVEALS AN OLIGOSACCHARIDE ACCEPTOR BINDING SITE
J.Mol.Biol., 318, 2002
|
|
1O0S
 
 | | Crystal Structure of Ascaris suum Malic Enzyme Complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, NAD-dependent malic enzyme, TARTRONATE | | Authors: | Rao, G.S, Coleman, D.E, Karsten, W.E, Cook, P.F, Harris, B.G. | | Deposit date: | 2003-02-24 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies on Ascaris suum NAD-malic enzyme bound to reduced cofactor and identification of an effector site.
J.Biol.Chem., 278, 2003
|
|
1O0T
 
 | | CRYSTAL STRUCTURE OF A COLD ADAPTED ALKALINE PROTEASE FROM PSEUDOMONAS TAC II 18, CO-CRYSTALLIZED WITH 5 mM EDTA (5 DAYS) | | Descriptor: | CALCIUM ION, SULFATE ION, serralysin | | Authors: | Ravaud, S, Gouet, P, Haser, R, Aghajari, N. | | Deposit date: | 2003-02-24 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the role of divalent metal ions in a bacterial psychrophilic metalloprotease: binding studies of an enzyme in the crystalline state by x-ray crystallography.
J.Bacteriol., 185, 2003
|
|
1O0V
 
 | | The crystal structure of IgE Fc reveals an asymmetrically bent conformation | | Descriptor: | GLYCEROL, Immunoglobulin heavy chain epsilon-1, SULFATE ION, ... | | Authors: | Wan, T, Beavil, R.L, Fabiane, S.M, Beavil, A.J, Sohi, M.K, Keown, M, Young, R.J, Henry, A.J, Owens, R.J, Gould, H.J, Sutton, B.J. | | Deposit date: | 2002-09-06 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of IgE Fc reveals an asymmetrically bent conformation
NAT.IMMUNOL., 3, 2002
|
|
1O0W
 
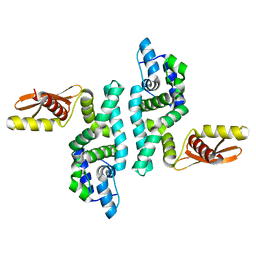 | |
1O0X
 
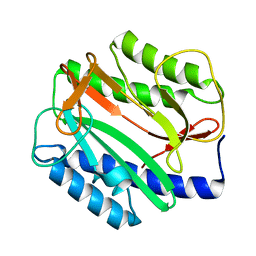 | |
1O12
 
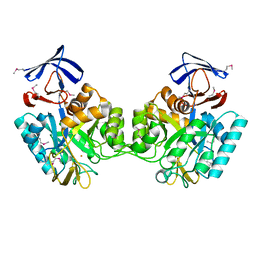 | |
1O13
 
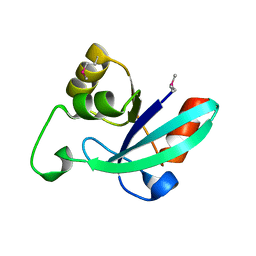 | |
1O15
 
 | |
1O16
 
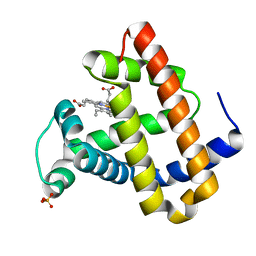 | | RECOMBINANT SPERM WHALE MYOGLOBIN H64D/V68S/D122N MUTANT (MET) | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Phillips Jr, G.N. | | Deposit date: | 2002-10-25 | | Release date: | 2003-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular engineering of myoglobin: influence of residue 68 on the rate and the
enantioselectivity of oxidation reactions catalyzed by H64D/V68X myoglobin
Biochemistry, 42, 2003
|
|
1O17
 
 | |
1O18
 
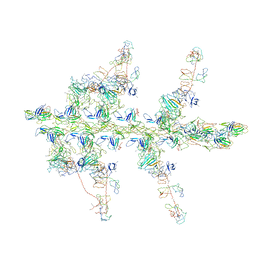 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-04 | | Release date: | 2002-11-27 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.Struct.Biol., 138, 2002
|
|
1O19
 
 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-15 | | Release date: | 2002-11-27 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.Struct.Biol., 138, 2002
|
|
1O1A
 
 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-18 | | Release date: | 2002-12-04 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.Struct.Biol., 138, 2002
|
|
1O1B
 
 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-15 | | Release date: | 2002-12-04 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.Struct.Biol., 138, 2002
|
|
1O1C
 
 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-18 | | Release date: | 2002-12-04 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.Struct.Biol., 138, 2002
|
|
1O1D
 
 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-18 | | Release date: | 2002-12-04 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.Struct.Biol., 138, 2002
|
|
