7TWA
 
 | | Crystal structure of apo BesC from Streptomyces cattleya | | Descriptor: | 1,3-BUTANEDIOL, 4-chloro-allylglycine synthase, ACETATE ION, ... | | Authors: | Neugebauer, M.E, McBride, M.J, Boal, A.K, Chang, M.C.Y. | | Deposit date: | 2022-02-07 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate-Triggered mu-Peroxodiiron(III) Intermediate in the 4-Chloro-l-Lysine-Fragmenting Heme-Oxygenase-like Diiron Oxidase (HDO) BesC: Substrate Dissociation from, and C4 Targeting by, the Intermediate.
Biochemistry, 61, 2022
|
|
7SA3
 
 | | Structure of a monomeric photosystem II core complex from a cyanobacterium acclimated to far-red light | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Gisriel, C.J, Bryant, D.A, Brudvig, G.W. | | Deposit date: | 2021-09-22 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | Structure of a monomeric photosystem II core complex from a cyanobacterium acclimated to far-red light reveals the functions of chlorophylls d and f.
J.Biol.Chem., 298, 2021
|
|
8R2I
 
 | | Cryo-EM Structure of native Photosystem II assembly intermediate from Chlamydomonas reinhardtii | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Fadeeva, M, Klaiman, D, Kandiah, E, Nelson, N. | | Deposit date: | 2023-11-06 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of native photosystem II assembly intermediate from Chlamydomonas reinhardtii .
Front Plant Sci, 14, 2023
|
|
6WY0
 
 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-40 A.K.A 7-[(1R)-1-phenyl-3-{[(1r,4r)-4-phenylcyclohexyl]amino}propyl]-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-{(1R)-1-phenyl-3-[(trans-4-phenylcyclohexyl)amino]propyl}-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, ... | | Authors: | Khan, J.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Bioorg.Med.Chem., 28, 2020
|
|
6WY5
 
 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-37 A.K.A 7-(1-phenyl-3-(((1S,3S)-3-phenyl-2,3-dihydro-1H-inden-1-yl)amino)propyl)-1H-[1,2,3]triazolo[4,5-b]pyridin-5-amine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(1R)-1-phenyl-3-{[(1S,3S)-3-phenyl-2,3-dihydro-1H-inden-1-yl]amino}propyl]-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, ... | | Authors: | Khan, J.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.898 Å) | | Cite: | Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Bioorg.Med.Chem., 28, 2020
|
|
7UT5
 
 | | Acinetobacter baumannii dihydroorotate dehydrogenase bound with inhibitor DSM186 | | Descriptor: | (4R)-7-methyl-N-[4-(pentafluoro-lambda~6~-sulfanyl)phenyl]imidazo[1,2-a]pyrimidin-5-amine, Dihydroorotate dehydrogenase (quinone), FLAVIN MONONUCLEOTIDE, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2022-04-26 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Repurposed dihydroorotate dehydrogenase inhibitors with efficacy against drug-resistant Acinetobacter baumannii.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5JRD
 
 | | E. coli Hydrogenase-1 variant P508A | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Phillips, S.E.V, Armstrong, F.A, Evans, R.M, Brooke, E.J, Islam, S.T.A. | | Deposit date: | 2016-05-06 | | Release date: | 2017-03-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Importance of the Active Site "Canopy" Residues in an O2-Tolerant [NiFe]-Hydrogenase.
Biochemistry, 56, 2017
|
|
8EKT
 
 | | CYP51 from Acanthamoeba castellanii in complex with the tetrazole-based IND inhibitor VT-1161(VT1) | | Descriptor: | (R)-2-(2,4-Difluorophenyl)-1,1-difluoro-3-(1H-tetrazol-1-yl)-1-(5-(4-(2,2,2-trifluoroethoxy)phenyl)pyridin-2-yl)propan-2-ol, PROTOPORPHYRIN IX CONTAINING FE, sterol 14a-demethylase | | Authors: | Hargrove, T.Y, Wawrzak, Z, Lepesheva, G.I. | | Deposit date: | 2022-09-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Identification of Potent and Selective Inhibitors of Acanthamoeba : Structural Insights into Sterol 14 alpha-Demethylase as a Key Drug Target.
J.Med.Chem., 67, 2024
|
|
4UDY
 
 | | NCO- bound to cluster C of Ni,Fe-CO dehydrogenase at true-atomic resolution | | Descriptor: | CARBON MONOXIDE DEHYDROGENASE 2, FE (II) ION, FE(3)-NI(1)-S(4) CLUSTER, ... | | Authors: | Fesseler, J, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2014-12-12 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | How the [Nife4 S4 ] Cluster of Co Dehydrogenase Activates Co2 and Nco(.)
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
9R3J
 
 | | Crystal structure of human MAO B in complex with (E)-3-(benzo[d][1,3]dioxol-5-yl)-1-(3-(trifluoromethyl)phenyl)prop-2-en-1-one (chalcone inhibitor, 4e) | | Descriptor: | (~{E})-3-(1,3-benzodioxol-5-yl)-1-[3-(trifluoromethyl)phenyl]prop-2-en-1-one, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Marchese, S, Binda, C. | | Deposit date: | 2025-05-05 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, synthesis, and biological evaluation of chalcone derivatives as selective Monoamine Oxidase-B inhibitors with potential neuroprotective effects.
Eur.J.Med.Chem., 298, 2025
|
|
6FWC
 
 | | Crystal structure of human monoamine oxidase B (MAO B) in complex with fluorophenyl-chromone-carboxamide | | Descriptor: | Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Reis, J, Manzella, N, Cagide, F, Mialet-Perez, J, Uriarte, E, Parini, A, Borges, F, Binda, C. | | Deposit date: | 2018-03-06 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tight-Binding Inhibition of Human Monoamine Oxidase B by Chromone Analogs: A Kinetic, Crystallographic, and Biological Analysis.
J. Med. Chem., 61, 2018
|
|
1N40
 
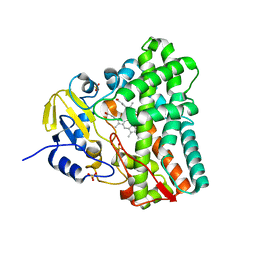 | | Atomic structure of CYP121, a mycobacterial P450 | | Descriptor: | Cytochrome P450 121, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Leys, D, Mowat, C.G, McLean, K.J, Richmond, A, Chapman, S.K, Walkinshaw, M.D, Munro, A.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-10-30 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Atomic structure of Mycobacterium tuberculosis CYP121 to 1.06 A reveals novel features of cytochrome P450.
J.Biol.Chem., 278, 2003
|
|
6D9Y
 
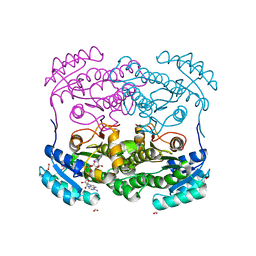 | |
9FXQ
 
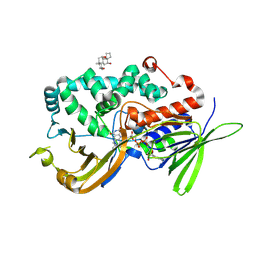 | | Ancestral Prenylcysteine Oxidase 1 (PCYOX1) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ancestral Prenylcysteine Oxidase 1 (PCYOX1), FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Barone, M, Mattevi, A. | | Deposit date: | 2024-07-02 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Evolution, structure, and drug-metabolizing activity of mammalian prenylcysteine oxidases.
J.Biol.Chem., 300, 2024
|
|
6FW0
 
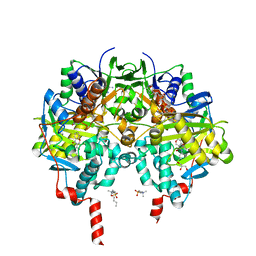 | | Crystal structure of human monoamine oxidase B (MAO B) in complex with chlorophenyl-chromone-carboxamide | | Descriptor: | Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Reis, J, Manzella, N, Cagide, F, Mialet-Perez, J, Uriarte, E, Parini, A, Borges, F, Binda, C. | | Deposit date: | 2018-03-05 | | Release date: | 2018-04-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tight-Binding Inhibition of Human Monoamine Oxidase B by Chromone Analogs: A Kinetic, Crystallographic, and Biological Analysis.
J. Med. Chem., 61, 2018
|
|
5KXJ
 
 | | Crystal Structure of L-Aspartate Oxidase from Salmonella typhimurium in the Complex with Substrate L-Aspartate | | Descriptor: | 1,2-ETHANEDIOL, ASPARTIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Osipiuk, J, Mulligan, R, Makowska-Grzyska, M, Maltseva, N, Shatsman, S, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-20 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure of L-Aspartate Oxidase from Salmonella typhimurium in the Complex with Substrate L-Aspartate
To Be Published
|
|
4UDX
 
 | | CO2 bound to cluster C of Ni,Fe-CO dehydrogenase at true-atomic resolution | | Descriptor: | CARBON DIOXIDE, CARBON MONOXIDE DEHYDROGENASE 2, FE (II) ION, ... | | Authors: | Fesseler, J, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2014-12-12 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | How the [NiFe4S4] Cluster of CO Dehydrogenase Activates CO2 and NCO(-).
Angew. Chem. Int. Ed. Engl., 54, 2015
|
|
6VI6
 
 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - a substrate monodentately bound structure | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VBY
 
 | | Cinnamate 4-hydroxylase (C4H1) from Sorghum bicolor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cinnamic acid 4-hydroxylase, GLYCEROL, ... | | Authors: | Zhang, B, Kang, C, Lewis, K.M. | | Deposit date: | 2019-12-19 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Function of the Cytochrome P450 Monooxygenase Cinnamate 4-hydroxylase fromSorghum bicolor.
Plant Physiol., 183, 2020
|
|
1BCC
 
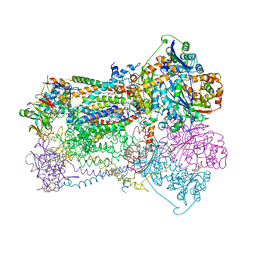 | | CYTOCHROME BC1 COMPLEX FROM CHICKEN | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.-I, Kim, K.K, Hung, L.-W, Crofts, A.R, Berry, E.A, Kim, S.-H. | | Deposit date: | 1998-03-23 | | Release date: | 1998-08-19 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Electron transfer by domain movement in cytochrome bc1.
Nature, 392, 1998
|
|
9CW3
 
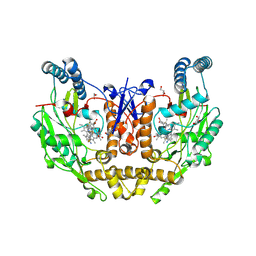 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain bound with 6-(5-((4,4-difluoropiperidin-1-yl)methyl)-2,3-difluorophenyl)-4-methylpyridin-2-amine dihydrochloride | | Descriptor: | (6M)-6-{5-[(4,4-difluoropiperidin-1-yl)methyl]-2,3-difluorophenyl}-4-methylpyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2024-07-29 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Truncated pyridinylbenzylamines: Potent, selective, and highly membrane permeable inhibitors of human neuronal nitric oxide synthase.
Bioorg.Med.Chem., 124, 2025
|
|
6Q9E
 
 | |
8CJL
 
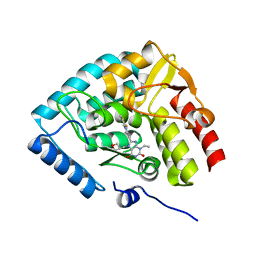 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor TPT-004 | | Descriptor: | 3-ethyl-8-[(2-methyl-5~{H}-imidazo[2,1-b][1,3]thiazol-6-yl)methyl]-7-(oxetan-3-ylmethyl)purine-2,6-dione, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure-Based Design of Xanthine-Imidazopyridines and -Imidazothiazoles as Highly Potent and In Vivo Efficacious Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
5F37
 
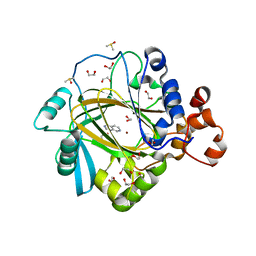 | | Crystal structure of human KDM4A in complex with compound 58 | | Descriptor: | 1,2-ETHANEDIOL, 3H-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.-V, Dempster, S, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F5A
 
 | | Crystal Structure of human JMJD2D complexed with KDOAM16 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(furan-2-ylmethylamino)methyl]pyridine-4-carboxylic acid, Lysine-specific demethylase 4D, ... | | Authors: | Krojer, T, Vollmar, M, Crawley, L, Bradley, A.R, Szykowska, A, Ruda, G.F, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
