3MXL
 
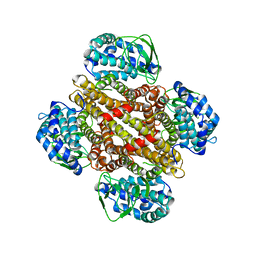 | |
3MUO
 
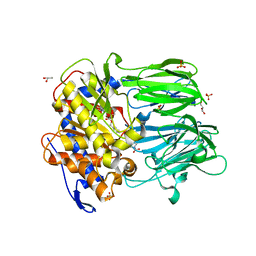 | | APPEP_PEPCLOSE+PP closed state | | Descriptor: | GLYCEROL, N-BENZYLOXYCARBONYL-L-PROLYL-L-PROLINAL, Prolyl endopeptidase, ... | | Authors: | Chiu, T.K. | | Deposit date: | 2010-05-03 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Route of Substrate Entry in Prolyl Endopeptidase
TO BE PUBLISHED
|
|
4B2T
 
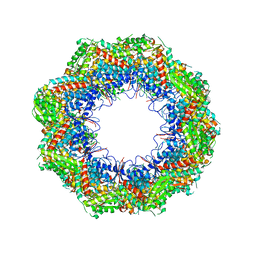 | | The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT ALPHA, T-COMPLEX PROTEIN 1 SUBUNIT BETA, T-COMPLEX PROTEIN 1 SUBUNIT DELTA, ... | | Authors: | Kalisman, N, Schroeder, G.F, Levitt, M. | | Deposit date: | 2012-07-17 | | Release date: | 2013-03-20 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | The Crystal Structures of the Eukaryotic Chaperonin Cct Reveal its Functional Partitioning
Structure, 21, 2013
|
|
4B7W
 
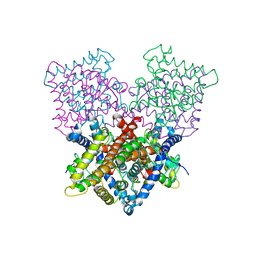 | | Ligand binding domain human hepatocyte nuclear factor 4alpha: Apo form | | Descriptor: | HEPATOCYTE NUCLEAR FACTOR 4-ALPHA | | Authors: | Dudasova, Z, Okvist, M, Kretova, M, Ondrovicova, G, Skrabana, R, LeGuevel, R, Salbert, G, Leonard, G, McSweeney, S, Barath, P. | | Deposit date: | 2012-08-24 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Fatty Acids are not Essential Structural Components of Hepatocyte Nuclear Factor 4Alpha
To be Published
|
|
3N71
 
 | |
4AIG
 
 | | ADAMALYSIN II WITH PHOSPHONATE INHIBITOR | | Descriptor: | ADAMALYSIN II, CALCIUM ION, N-[(FURAN-2-YL)CARBONYL]-(S)-LEUCYL-(R)-[1-AMINO-2(1H-INDOL-3-YL)ETHYL]-PHOSPHONIC ACID, ... | | Authors: | Pochetti, G, Mazza, F, Gavuzzo, E, Cirilli, M. | | Deposit date: | 1997-10-16 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2 angstrom X-ray structure of adamalysin II complexed with a peptide phosphonate inhibitor adopting a retro-binding mode.
FEBS Lett., 418, 1997
|
|
4ANL
 
 | |
406D
 
 | | 5'-R(*CP*AP*CP*CP*GP*GP*AP*UP*GP*GP*UP*(BRO) UP*CP*GP*GP*UP*G)-3' | | Descriptor: | RNA (5'-R(*CP*AP*CP*CP*GP*GP*AP*UP*GP*GP*UP*(5BU)P*CP*GP*GP*UP*G)-3') | | Authors: | Shah, S.A, Brunger, A.T. | | Deposit date: | 1998-06-22 | | Release date: | 1999-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A crystal structure of a statically disordered 17 base-pair RNA duplex: principles of RNA crystal packing and its effect on nucleic acid structure.
J.Mol.Biol., 285, 1999
|
|
411D
 
 | | DUPLEX [5'-D(GCGTA+TACGC)]2 WITH INCORPORATED 2'-O-METHOXYETHYL RIBONUCLEOSIDE | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(T39)P*AP*CP*GP*C)-3') | | Authors: | Tereshko, V, Portmann, S, Tay, E.C, Martin, P, Natt, F, Altmann, K.H, Egli, M. | | Deposit date: | 1998-06-30 | | Release date: | 1998-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Correlating structure and stability of DNA duplexes with incorporated 2'-O-modified RNA analogues.
Biochemistry, 37, 1998
|
|
419D
 
 | |
421P
 
 | | THREE-DIMENSIONAL STRUCTURES OF H-RAS P21 MUTANTS: MOLECULAR BASIS FOR THEIR INABILITY TO FUNCTION AS SIGNAL SWITCH MOLECULES | | Descriptor: | H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Krengel, U, John, J, Scherer, A, Kabsch, W, Wittinghofer, A, Pai, E.F. | | Deposit date: | 1991-06-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules.
Cell(Cambridge,Mass.), 62, 1990
|
|
441D
 
 | |
449D
 
 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', BENZIMIDAZOLE DERIVATIVE COMPLEX | | Descriptor: | 2'-(3-IODOPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Squire, C.J, Baker, L.J, Clark, G.R, Martin, R.F, White, J. | | Deposit date: | 1999-01-20 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
3N6R
 
 | | CRYSTAL STRUCTURE OF the holoenzyme of PROPIONYL-COA CARBOXYLASE (PCC) | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Propionyl-CoA carboxylase, alpha subunit, ... | | Authors: | Huang, C.S, Sadre-Bazzaz, K, Tong, L. | | Deposit date: | 2010-05-26 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the alpha(6)beta(6) holoenzyme of propionyl-coenzyme A carboxylase.
Nature, 466, 2010
|
|
488D
 
 | | CATALYTIC RNA ENZYME-PRODUCT COMPLEX | | Descriptor: | CADMIUM ION, FIRST RNA FRAGMENT OF CLEAVED SUBSTRATE, RNA RIBOZYME STRAND, ... | | Authors: | Murray, J.B, Szoke, H, Szoke, A, Scott, W.G. | | Deposit date: | 2000-02-25 | | Release date: | 2000-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Capture and visualization of a catalytic RNA enzyme-product complex using crystal lattice trapping and X-ray holographic reconstruction.
Mol.Cell, 5, 2000
|
|
4A7J
 
 | | Symmetric Dimethylation of H3 Arginine 2 is a Novel Histone Mark that Supports Euchromatin Maintenance | | Descriptor: | HISTONE H3.1T, WD REPEAT-CONTAINING PROTEIN 5 | | Authors: | Migliori, V, Muller, J, Phalke, S, Low, D, Bezzi, M, ChuenMok, W, Gunaratne, J, Capasso, P, Bassi, C, Cecatiello, V, DeMarco, A, Blackstock, W, Kuznetsov, V, Amati, B, Mapelli, M, Guccione, E. | | Deposit date: | 2011-11-14 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Symmetric Dimethylation of H3R2 is a Newly Identified Histone Mark that Supports Euchromatin Maintenance
Nat.Struct.Mol.Biol., 19, 2012
|
|
3N35
 
 | | Erythrina corallodendron lectin mutant (Y106G) with N-Acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, Lectin, ... | | Authors: | Thamotharan, S, Karthikeyan, T, Kulkarni, K.A, Shetty, K.N, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2010-05-19 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modification of the sugar specificity of a plant lectin: structural studies on a point mutant of Erythrina corallodendron lectin.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2VV3
 
 | | hPPARgamma Ligand binding domain in complex with 4-oxoDHA | | Descriptor: | (6E,10Z,13Z,16Z,19Z)-4-oxodocosa-6,10,13,16,19-pentaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Activation of Ppargamma by Oxidized Fatty Acids.
Nat.Struct.Mol.Biol., 15, 2008
|
|
4A39
 
 | | Metallo-carboxypeptidase from Pseudomonas Aeruginosa in complex with (2-guanidinoethylmercapto)succinic acid | | Descriptor: | (2-GUANIDINOETHYLMERCAPTO)SUCCINIC ACID, METALLO-CARBOXYPEPTIDASE, ZINC ION | | Authors: | Otero, A, Rodriguez de la Vega, M, Tanco, S.M, Lorenzo, J, Aviles, F.X, Reverter, D. | | Deposit date: | 2011-09-30 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Novel Structure of a Cytosolic M14 Metallocarboxypeptidase (Ccp) from Pseudomonas Aeruginosa: A Model for Mammalian Ccps.
Faseb J., 26, 2012
|
|
4AKF
 
 | |
3NEN
 
 | |
4AKR
 
 | | Crystal Structure of the cytoplasmic actin capping protein Cap32_34 from Dictyostelium discoideum | | Descriptor: | F-ACTIN-CAPPING PROTEIN SUBUNIT ALPHA, F-ACTIN-CAPPING PROTEIN SUBUNIT BETA | | Authors: | Eckert, C, Goretzki, A, Faberova, M, Kollmar, M. | | Deposit date: | 2012-02-28 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conservation and Divergence between Cytoplasmic and Muscle-Specific Actin Capping Proteins: Insights from the Crystal Structure of Cytoplasmic CAP32/34 from Dictyostelium Discoideum.
Bmc Struct.Biol., 12, 2012
|
|
4AI9
 
 | | JMJD2A Complexed with Daminozide | | Descriptor: | CHLORIDE ION, DAMINOZIDE, GLYCEROL, ... | | Authors: | Chowdhury, R, Schofield, C.J. | | Deposit date: | 2012-02-08 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Plant growth regulator daminozide is a selective inhibitor of human KDM2/7 histone demethylases.
J. Med. Chem., 55, 2012
|
|
3NUL
 
 | | Profilin I from Arabidopsis thaliana | | Descriptor: | GLYCEROL, PROFILIN I, SULFATE ION | | Authors: | Thorn, K, Christensen, H.E.M, Shigeta, R, Huddler, D, Chua, N.-H, Shalaby, L, Lindberg, U, Schutt, C.E. | | Deposit date: | 1996-11-27 | | Release date: | 1997-12-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of a major allergen from plants.
Structure, 5, 1997
|
|
4AMK
 
 | |
