8GJ8
 
 | | RAD51C C-terminal domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAD51C | | Authors: | Arvai, A.S, Tainer, J.A, Williams, G. | | Deposit date: | 2023-03-15 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles.
Nat Commun, 14, 2023
|
|
6OBE
 
 | | Ricin A chain bound to VHH antibody V6H8 | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, NICKEL (II) ION, ... | | Authors: | Rudolph, M.J. | | Deposit date: | 2019-03-20 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | Intracellular Neutralization of Ricin Toxin by Single-domain Antibodies Targeting the Active Site.
J.Mol.Biol., 432, 2020
|
|
6OBO
 
 | | Ricin A chain bound to VHH antibody V6A6 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Rudolph, M.J. | | Deposit date: | 2019-03-21 | | Release date: | 2020-04-01 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Intracellular Neutralization of Ricin Toxin by Single-domain Antibodies Targeting the Active Site.
J.Mol.Biol., 432, 2020
|
|
6Q7O
 
 | | Crystal structure of OE1 | | Descriptor: | CALCIUM ION, OE1 | | Authors: | Levy, C.W. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and evolution of an enzyme with a non-canonical organocatalytic mechanism.
Nature, 570, 2019
|
|
6PZ0
 
 | | Crystal structure of oxidized iodotyrosine deiodinase (IYD) bound to FMN and L-Tyrosine | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, TYROSINE, ... | | Authors: | Sun, Z, Kavran, J.M, Rokita, S.E. | | Deposit date: | 2019-07-31 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The minimal structure for iodotyrosine deiodinase function is defined by an outlier protein from the thermophilic bacterium Thermotoga neapolitana.
J.Biol.Chem., 297, 2021
|
|
8FUL
 
 | |
8FUM
 
 | | AibH1H2 metalated with Fe in the presence of Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amidohydrolase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Powell, M.M, Rittle, J. | | Deposit date: | 2023-01-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Enzymatic Hydroxylation of Aliphatic C-H Bonds by a Mn/Fe Cofactor.
J.Am.Chem.Soc., 145, 2023
|
|
6Q7N
 
 | |
8FUO
 
 | | Fe-bound AibH1H2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amidohydrolase, FE (III) ION | | Authors: | Powell, M.M, Rittle, J. | | Deposit date: | 2023-01-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Enzymatic Hydroxylation of Aliphatic C-H Bonds by a Mn/Fe Cofactor.
J.Am.Chem.Soc., 145, 2023
|
|
6Q7L
 
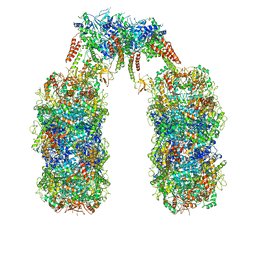 | | Spiral structure of E. coli RavA in the RavA-LdcI cage-like complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase RavA, Inducible lysine decarboxylase, ... | | Authors: | Arragain, B, Felix, J, Malet, H, Gutsche, I, Jessop, M. | | Deposit date: | 2018-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural insights into ATP hydrolysis by the MoxR ATPase RavA and the LdcI-RavA cage-like complex.
Commun Biol, 3, 2020
|
|
8FUN
 
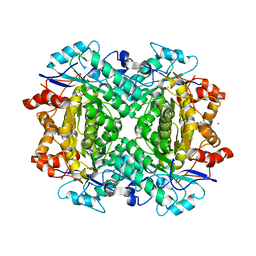 | | Enzymatically Active, Mn/Fe Metallated Form of AibH1H2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amidohydrolase, FE (III) ION, ... | | Authors: | Powell, M.M, Rittle, J. | | Deposit date: | 2023-01-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Enzymatic Hydroxylation of Aliphatic C-H Bonds by a Mn/Fe Cofactor.
J.Am.Chem.Soc., 145, 2023
|
|
6QAF
 
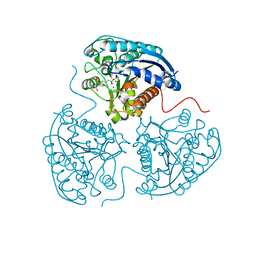 | | Crystal structure of human Arginase-1 at pH 9.0 in complex with CB-1158/INCB001158 | | Descriptor: | Arginase-1, MANGANESE (II) ION, SODIUM ION, ... | | Authors: | Grobben, Y, Uitdehaag, J.C.M, Tabak, W.W.A, Zaman, G.J.R. | | Deposit date: | 2018-12-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into human Arginase-1 pH dependence and its inhibition by the small molecule inhibitor CB-1158.
J Struct Biol X, 4, 2020
|
|
2WEU
 
 | |
6OAJ
 
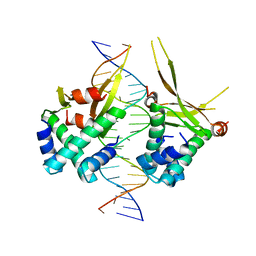 | | HUaE34K 19bp SYM DNA | | Descriptor: | DNA (5'-D(P*CP*GP*GP*TP*TP*CP*AP*AP*TP*TP*GP*GP*CP*AP*CP*GP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*CP*GP*TP*GP*CP*CP*AP*AP*TP*TP*GP*AP*AP*CP*CP*GP*C)-3'), DNA-binding protein HU-alpha | | Authors: | Remesh, S.G, Hammel, M. | | Deposit date: | 2019-03-16 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.092 Å) | | Cite: | Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun, 11, 2020
|
|
8GF3
 
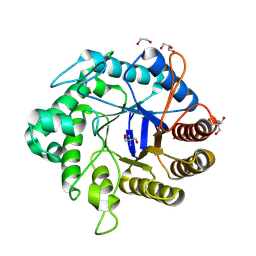 | | Crystallographic structure from BlMan5_7 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GH5 Mannanase, ... | | Authors: | Briganti, L, Araujo, E.A, Polikarpov, I. | | Deposit date: | 2023-03-07 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Unravelling biochemical and structural features of Bacillus licheniformis GH5 mannanase using site-directed mutagenesis and high-resolution protein crystallography studies.
Int.J.Biol.Macromol., 274, 2024
|
|
8G6D
 
 | |
6OBM
 
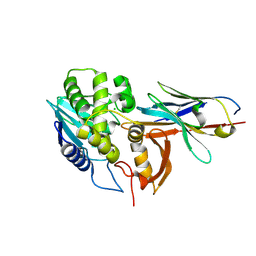 | | Ricin A chain bound to VHH antibody V6A7 | | Descriptor: | CHLORIDE ION, Ricin A chain, VHH antibody V6A7 | | Authors: | Rudolph, M.J. | | Deposit date: | 2019-03-21 | | Release date: | 2020-04-01 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Intracellular Neutralization of Ricin Toxin by Single-domain Antibodies Targeting the Active Site.
J.Mol.Biol., 432, 2020
|
|
6OQE
 
 | |
6QD0
 
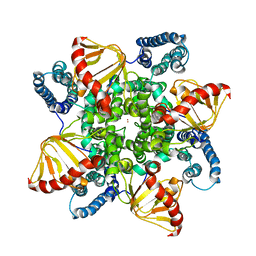 | | MloK1 model from single particle analysis of 2D crystals, class 4 (compact/open conformation) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Righetto, R, Biyani, N, Kowal, J, Chami, M, Stahlberg, H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving high-resolution information from disordered 2D crystals by single-particle cryo-EM.
Nat Commun, 10, 2019
|
|
6PWA
 
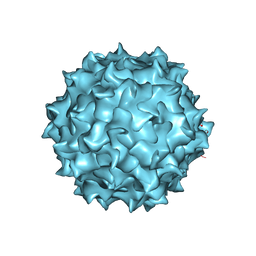 | | AAV8 human HEK293-produced, full capsid | | Descriptor: | Capsid protein | | Authors: | Paulk, N.K, Poweleit, N. | | Deposit date: | 2019-07-22 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Methods Matter: Standard Production Platforms for Recombinant AAV Produce Chemically and Functionally Distinct Vectors.
Mol Ther Methods Clin Dev, 18, 2020
|
|
6QCZ
 
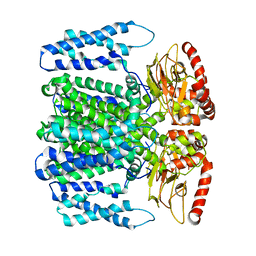 | | MloK1 model from single particle analysis of 2D crystals, class 3 (intermediate extended conformation) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Righetto, R, Biyani, N, Kowal, J, Chami, M, Stahlberg, H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Retrieving high-resolution information from disordered 2D crystals by single-particle cryo-EM.
Nat Commun, 10, 2019
|
|
6QD3
 
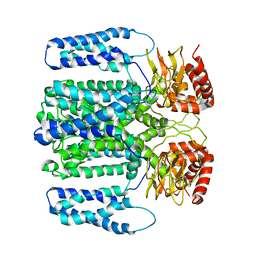 | | MloK1 model from single particle analysis of 2D crystals, class 7 (intermediate conformation) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Righetto, R, Biyani, N, Kowal, J, Chami, M, Stahlberg, H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Retrieving high-resolution information from disordered 2D crystals by single-particle cryo-EM.
Nat Commun, 10, 2019
|
|
6QDI
 
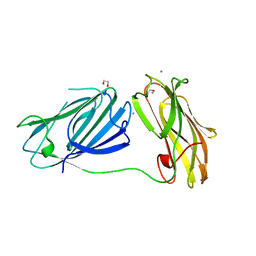 | | anti-sigma factor domain-containing protein from Clostridium clariflavum | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PA14 domain-containing protein | | Authors: | Voronov, M, Bayer, E.A, Livnah, O. | | Deposit date: | 2019-01-01 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Distinctive ligand-binding specificities of tandem PA14 biomass-sensory elements from Clostridium thermocellum and Clostridium clariflavum.
Proteins, 87, 2019
|
|
6QFW
 
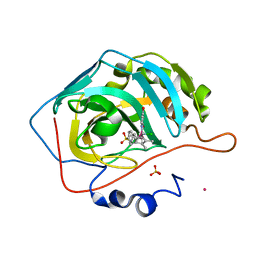 | | Human carbonic anhydrase II with bound IrCp* complex (cofactor 9) to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 4-[3-(8-chloranyl-2',3',4',5',6'-pentamethyl-4-oxidanyl-7-oxidanylidene-spiro[1$l^{4},8$l^{4}-diaza-9$l^{7}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{7}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)propyl]benzenesulfonamide, Carbonic anhydrase 2, IRIDIUM ION, ... | | Authors: | Rebelein, J.G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Chemical Optimization of Whole-Cell Transfer Hydrogenation Using Carbonic Anhydrase as Host Protein.
Acs Catalysis, 9, 2019
|
|
6QFV
 
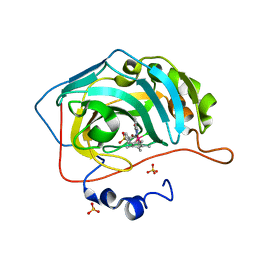 | | Human carbonic anhydrase II with bound IrCp* complex (cofactor 8) to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 4-[2-(9-chloranyl-2',3',4',5',6'-pentamethyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1(6),2,4-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, SULFATE ION, ... | | Authors: | Rebelein, J.G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Chemical Optimization of Whole-Cell Transfer Hydrogenation Using Carbonic Anhydrase as Host Protein.
Acs Catalysis, 9, 2019
|
|
