1N7V
 
 | | THE RECEPTOR-BINDING PROTEIN P2 OF BACTERIOPHAGE PRD1: CRYSTAL FORM III | | Descriptor: | ACETATE ION, Adsorption protein P2, CALCIUM ION | | Authors: | Xu, L, Benson, S.D, Butcher, S.J, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2002-11-18 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Receptor Binding Protein P2 of PRD1, a
Virus Targeting Antibiotic-Resistant Bacteria,
Has a Novel Fold Suggesting Multiple Functions.
Structure, 11, 2003
|
|
1N7W
 
 | | Crystal Structure of Human Serum Transferrin, N-Lobe L66W mutant | | Descriptor: | CARBONATE ION, FE (III) ION, Serotransferrin | | Authors: | Adams, T.E, Mason, A.B, He, Q.Y, Halbrooks, P.J, Briggs, S.K, Smith, V.C, MacGillivray, R.T, Everse, S.J. | | Deposit date: | 2002-11-18 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Position of Arginine 124 Controls the Rate of Iron Release from the N-lobe of Human Serum Transferrin. A Structural Study
J.Biol.Chem., 278, 2003
|
|
1N7X
 
 | | HUMAN SERUM TRANSFERRIN, N-LOBE Y45E MUTANT | | Descriptor: | CARBONATE ION, FE (III) ION, Serotransferrin | | Authors: | Adams, T.E, Mason, A.B, He, Q.Y, Halbrooks, P.J, Briggs, S.K, Smith, V.C, Macgillivray, R.T, Everse, S.J. | | Deposit date: | 2002-11-18 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | THE POSITION OF ARGININE 124 CONTROLS THE RATE OF IRON RELEASE FROM THE N-LOBE OF HUMAN SERUM TRANSFERRIN. A STRUCTURAL STUDY
J.Biol.Chem., 278, 2003
|
|
1N7Y
 
 | | STREPTAVIDIN MUTANT N23E AT 1.96A | | Descriptor: | Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-11-18 | | Release date: | 2003-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1N7Z
 
 | | Structure and location of gene product 8 in the bacteriophage T4 baseplate | | Descriptor: | CHLORIDE ION, baseplate structural protein gp8 | | Authors: | Leiman, P.G, Shneider, M.M, Kostyuchenko, V.A, Chipman, P.R, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2002-11-18 | | Release date: | 2003-06-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and location of gene product 8 in the bacteriophage T4 baseplate
J.Mol.Biol., 328, 2003
|
|
1N80
 
 | | Bacteriophage T4 baseplate structural protein gp8 | | Descriptor: | CHLORIDE ION, baseplate structural protein gp8 | | Authors: | Leiman, P.G, Shneider, M.M, Kostyuchenko, V.A, Chipman, P.R, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2002-11-18 | | Release date: | 2003-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure and location of gene product 8 in the bacteriophage T4 baseplate
J.Mol.Biol., 328, 2003
|
|
1N81
 
 | |
1N82
 
 | | The high-resolution crystal structure of IXT6, a thermophilic, intracellular xylanase from G. stearothermophilus | | Descriptor: | GLYCEROL, SODIUM ION, intra-cellular xylanase | | Authors: | Solomon, V, Teplitsky, A, Golan, G, Gilboa, R, Reiland, V, Shulami, S, Moryles, S, Zolotnitsky, G, Shoham, Y, Shoham, G. | | Deposit date: | 2002-11-19 | | Release date: | 2003-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The high-resolution crystal structure of IXT6,
a thermophilic, intracellular xylanase from G. stearothermophilus
To be Published
|
|
1N83
 
 | | Crystal Structure of the complex between the Orphan Nuclear Hormone Receptor ROR(alpha)-LBD and Cholesterol | | Descriptor: | CHOLESTEROL, Nuclear receptor ROR-alpha | | Authors: | Kallen, J.A, Schlaeppi, J.M, Bitsch, F, Geisse, S, Geiser, M, Delhon, I, Fournier, B. | | Deposit date: | 2002-11-19 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray Structure of hROR(alpha) LBD at 1.63A: Structural and Functional data that Cholesterol or a Cholesterol derivative is the natural ligand of ROR(alpha)
Structure, 10, 2002
|
|
1N84
 
 | | HUMAN SERUM TRANSFERRIN, N-LOBE | | Descriptor: | CARBONATE ION, FE (III) ION, Serotransferrin | | Authors: | Adams, T.E, Mason, A.B, He, Q.Y, Halbrooks, P.J, Briggs, S.K, Smith, V.C, Macgillivray, R.T, Everse, S.J. | | Deposit date: | 2002-11-19 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | THE POSITION OF ARGININE 124 CONTROLS THE RATE OF IRON RELEASE FROM THE N-LOBE OF HUMAN SERUM TRANSFERRIN. A STRUCTURAL STUDY
J.Biol.Chem., 278, 2003
|
|
1N86
 
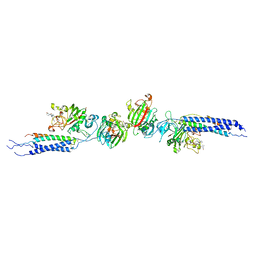 | | Crystal structure of human D-dimer from cross-linked fibrin complexed with GPR and GHRPLDK peptide ligands. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, CALCIUM ION, Fibrin alpha/alpha-E chain, ... | | Authors: | Yang, Z, Pandi, L, Doolittle, R.F. | | Deposit date: | 2002-11-19 | | Release date: | 2003-01-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of fragment double-D from cross-linked lamprey fibrin reveals isopeptide linkages across an unexpected D-D interface.
Biochemistry, 41, 2002
|
|
1N87
 
 | |
1N88
 
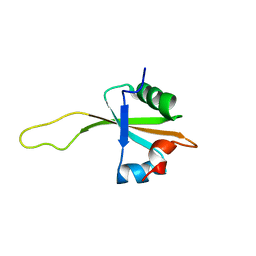 | | NMR structure of the ribosomal protein L23 from Thermus thermophilus. | | Descriptor: | Ribosomal protein L23 | | Authors: | Ohman, A, Rak, A, Dontsova, M, Garber, M.B, Hard, T. | | Deposit date: | 2002-11-20 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the ribosomal protein L23 from Thermus thermophilus.
J.Biomol.NMR, 26, 2003
|
|
1N89
 
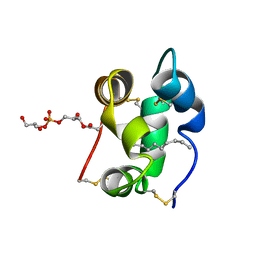 | | Solution structure of a liganded type 2 wheat non-specific Lipid Transfer Protein | | Descriptor: | 1-MYRISTOYL-2-HYDROXY-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL)], lipid transfer protein | | Authors: | Pons, J.L, de Lamotte, F, Gautier, M.F, Delsuc, M.A. | | Deposit date: | 2002-11-20 | | Release date: | 2003-03-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of a liganded type 2 wheat nonspecific lipid transfer protein.
J.Biol.Chem., 278, 2003
|
|
1N8B
 
 | | Bacteriophage T4 baseplate structural protein gp8 | | Descriptor: | BROMIDE ION, baseplate structural protein gp8 | | Authors: | Leiman, P.G, Shneider, M.M, Kostyuchenko, V.A, Chipman, P.R, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2002-11-20 | | Release date: | 2003-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and location of gene product 8 in the bacteriophage T4 baseplate
J.Mol.Biol., 328, 2003
|
|
1N8C
 
 | | Solution Structure of a Cis-Opened (10R)-N6-Deoxyadenosine Adduct of (9S,10R)-(9,10)-Epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene in a DNA Duplex | | Descriptor: | (9S,10R)-9-HYDROXY-7,8,9,10-TETRAHYDROBENZO[A]PYRENE, 5'-D(*CP*CP*TP*CP*GP*TP*GP*AP*CP*CP*G)-3', 5'-D(*CP*GP*GP*TP*CP*AP*CP*GP*AP*GP*G)-3' | | Authors: | Volk, D.E, Thiviyanathan, V, Rice, J.S, Luxon, B.A, Shah, J.H, Yagi, H, Sayer, J.M, Yeh, H.J.C, Jerina, D.M, Gorenstein, D.G. | | Deposit date: | 2002-11-20 | | Release date: | 2003-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Cis-Opened (10R)-N6-Deoxyadenosine Adduct of (9S,10R)-(9,10)-Epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene in a DNA Duplex
Biochemistry, 42, 2003
|
|
1N8E
 
 | | Fragment Double-D from Human Fibrin | | Descriptor: | Fibrin alpha/alpha-E chain, Fibrin beta chain, Fibrin gamma chain | | Authors: | Yang, Z, Pandi, L, Doolittle, R.F. | | Deposit date: | 2002-11-20 | | Release date: | 2003-01-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | The Crystal Structure of Fragment Double-D from Cross-Linked Lamprey Fibrin Reveals Isopeptide Linkages across an Unexpected D-D Interface
Biochemistry, 41, 2002
|
|
1N8F
 
 | | Crystal structure of E24Q mutant of phenylalanine-regulated 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHP synthase) from Escherichia Coli in complex with Mn2+ and PEP | | Descriptor: | DAHP Synthetase, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Shumilin, I.A, Bauerle, R, Kretsinger, R.H. | | Deposit date: | 2002-11-20 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The High-Resolution Structure of 3-Deoxy-D-arabino-heptulosonate-7-phosphate
Synthase Reveals a Twist in the Plane of Bound Phosphoenolpyruvate
Biochemistry, 42, 2003
|
|
1N8I
 
 | | Biochemical and Structural Studies of Malate Synthase from Mycobacterium tuberculosis | | Descriptor: | GLYOXYLIC ACID, MAGNESIUM ION, Probable malate synthase G | | Authors: | Smith, C.V, Huang, C.C, Miczak, A, Russell, D.G, Sacchettini, J.C, Honer zu Bentrup, K, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-11-20 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and structural studies of malate synthase from Mycobacterium tuberculosis
J.Biol.Chem., 278, 2003
|
|
1N8J
 
 | |
1N8K
 
 | |
1N8M
 
 | | Solution structure of Pi4, a four disulfide bridged scorpion toxin active on potassium channels | | Descriptor: | Potassium channel blocking toxin 4 | | Authors: | Guijarro, J.I, M'Barek, S, Olamendi-Portugal, T, Gomez-Lagunas, F, Garnier, D, Rochat, H, Possani, L.D, Sabatier, J.M, Delepierre, M. | | Deposit date: | 2002-11-21 | | Release date: | 2003-09-02 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Pi4, a short four-disulfide-bridged scorpion toxin specific of potassium channels.
Protein Sci., 12, 2003
|
|
1N8N
 
 | | Crystal structure of the Au3+ complex of AphA class B acid phosphatase/phosphotransferase from E. coli at 1.69 A resolution | | Descriptor: | Class B acid phosphatase, GOLD 3+ ION | | Authors: | Calderone, V, Forleo, C, Benvenuti, M, Rossolini, G.M, Thaller, M.C, Mangani, S. | | Deposit date: | 2002-11-21 | | Release date: | 2004-02-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The first structure of a bacterial class B Acid phosphatase reveals further structural heterogeneity among phosphatases of the haloacid dehalogenase fold.
J.Mol.Biol., 335, 2004
|
|
1N8O
 
 | |
1N8P
 
 | | Crystal Structure of cystathionine gamma-lyase from yeast | | Descriptor: | Cystathionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Messerschmidt, A, Worbs, M, Steegborn, C, Wahl, M.C, Huber, R, Clausen, T. | | Deposit date: | 2002-11-21 | | Release date: | 2002-12-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Determinants of Enzymatic Specificity in the Cys-Met-Metabolism PLP-Dependent Enzymes Family: Crystal Structure of Cystathionine gamma-lyase from Yeast and Intrafamiliar Structural Comparison
BIOL.CHEM., 384, 2003
|
|
