1TUG
 
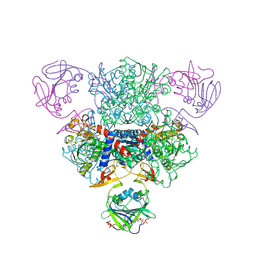 | | Aspartate Transcarbamoylase Catalytic Chain Mutant E50A Complex with Phosphonoacetamide, Malonate, and Cytidine-5-Prime-Triphosphate (CTP) | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Stieglitz, K, Stec, B, Baker, D.P, Kantrowitz, E.R. | | Deposit date: | 2004-06-24 | | Release date: | 2004-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Monitoring the Transition from the T to the R State in E.coli Aspartate Transcarbamoylase by X-ray Crystallography: Crystal Structures of the E50A Mutant Enzyme in Four Distinct Allosteric States.
J.Mol.Biol., 341, 2004
|
|
8BI8
 
 | | Structure of a cyclic beta-hairpin peptide derived from neuronal nitric oxide synthase | | Descriptor: | Nitric oxide synthase, brain | | Authors: | Balboa, J.R, Ostergaard, S, Stromgaard, K, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-01 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Development of a Potent Cyclic Peptide Inhibitor of the nNOS/PSD-95 Interaction.
J.Med.Chem., 66, 2023
|
|
4DJ5
 
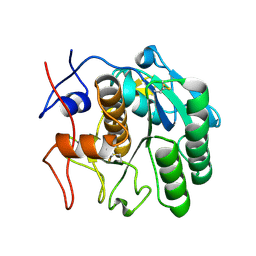 | |
1U8B
 
 | | Crystal structure of the methylated N-ADA/DNA complex | | Descriptor: | 5'-D(*AP*AP*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*T)-3', 5'-D(*TP*AP*AP*AP*TP*T)-3', 5'-D(P*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*AP*T)-3', ... | | Authors: | He, C, Hus, J.-C, Sun, L.J, Zhou, P, Norman, D.P.G, Dotsch, V, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | Deposit date: | 2004-08-05 | | Release date: | 2005-10-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A methylation-dependent electrostatic switch controls DNA repair and transcriptional activation by E. coli ada.
Mol.Cell, 20, 2005
|
|
8BFI
 
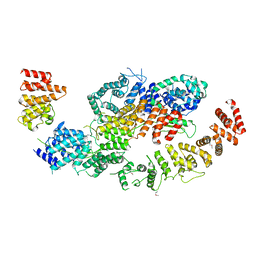 | | human CNOT1-CNOT10-CNOT11 module | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 10, CCR4-NOT transcription complex subunit 11 | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The human CNOT1-CNOT10-CNOT11 complex forms a structural platform for protein-protein interactions.
Cell Rep, 42, 2023
|
|
1U8L
 
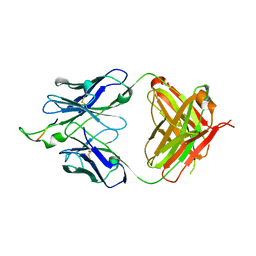 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide DLDRWAS | | Descriptor: | ANTIBODY 2F5 (HEAVY CHAIN), ANTIBODY 2F5 (LIGHT CHAIN), GP41 PEPTIDE | | Authors: | Bryson, S, Julien, J.-P, Hynes, R.C, Pai, E.F. | | Deposit date: | 2004-08-06 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications
J.Virol., 83, 2009
|
|
8BD8
 
 | | Crystal structure of TRIM33 alpha PHD-Bromo domain in complex with 8 | | Descriptor: | 1,3-dimethylbenzimidazol-2-one, CALCIUM ION, E3 ubiquitin-protein ligase TRIM33, ... | | Authors: | Tassone, G, Pozzi, C, Palomba, T. | | Deposit date: | 2022-10-18 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Exploiting ELIOT for Scaffold-Repurposing Opportunities: TRIM33 a Possible Novel E3 Ligase to Expand the Toolbox for PROTAC Design.
Int J Mol Sci, 23, 2022
|
|
1U8W
 
 | | Crystal structure of Arabidopsis thaliana nucleoside diphosphate kinase 1 | | Descriptor: | Nucleoside diphosphate kinase I | | Authors: | Im, Y.J, Kim, J.-I, Shen, Y, Na, Y, Han, Y.-J, Kim, S.-H, Song, P.-S, Eom, S.H. | | Deposit date: | 2004-08-07 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of Arabidopsis thaliana nucleoside diphosphate kinase-2 for phytochrome-mediated light signaling
J.Mol.Biol., 343, 2004
|
|
8AYZ
 
 | | Poliovirus type 2 (strain MEF-1) virus-like particle in complex with capsid binder compound 17 | | Descriptor: | 4-[[4-[1,3-bis(oxidanylidene)isoindol-2-yl]phenyl]sulfonylamino]benzoic acid, Capsid protein, VP0, ... | | Authors: | Bahar, M.W, Fry, E.E, Stuart, D.I. | | Deposit date: | 2022-09-04 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (1.88 Å) | | Cite: | A conserved glutathione binding site in poliovirus is a target for antivirals and vaccine stabilisation.
Commun Biol, 5, 2022
|
|
8B0B
 
 | | Crystal structure of human heparanase in complex with covalent inhibitor VB151 | | Descriptor: | (1~{S},2~{R},3~{R},4~{S},6~{S})-2-(2-acetamidoethoxy)-3,4,6-tris(oxidanyl)cyclohexane-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Armstrong, Z, Davies, G.J. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 4-O-Substituted Glucuronic Cyclophellitols are Selective Mechanism-Based Heparanase Inhibitors.
Chemmedchem, 18, 2023
|
|
1U9A
 
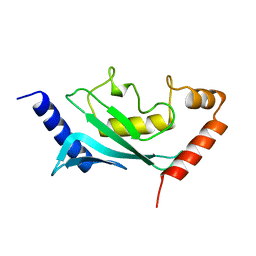 | | HUMAN UBIQUITIN-CONJUGATING ENZYME UBC9 | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME | | Authors: | Tong, H, Hateboer, G, Perrakis, A, Bernards, R, Sixma, T.K. | | Deposit date: | 1997-02-11 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of murine/human Ubc9 provides insight into the variability of the ubiquitin-conjugating system.
J.Biol.Chem., 272, 1997
|
|
8B8X
 
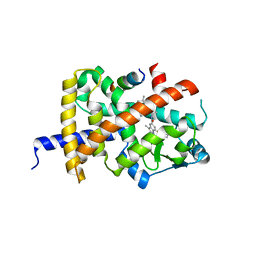 | | Crystal structure of PPARG and NCOR2 with SR10221, an inverse agonist | | Descriptor: | (2S)-2-{5-[(5-{[(1S)-1-(4-tert-butylphenyl)ethyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]-2-chlorophenoxy}propanoic acid, Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Siegel, S, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery and characterization of orally bioavailable 4-chloro-6-fluoroisophthalamides as covalent PPARG inverse-agonists.
Bioorg.Med.Chem., 78, 2022
|
|
8BD5
 
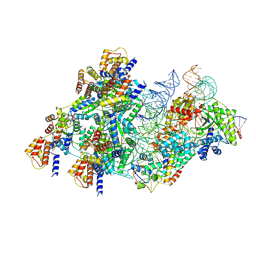 | | Cas12k-sgRNA-dsDNA-S15-TniQ-TnsC transposon recruitment complex | | Descriptor: | 30S ribosomal protein S15, ADENOSINE-5'-TRIPHOSPHATE, DNA non-target strand, ... | | Authors: | Schmitz, M, Querques, I, Oberli, S, Chanez, C, Jinek, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the assembly of the type V CRISPR-associated transposon complex.
Cell, 185, 2022
|
|
1U9J
 
 | |
1U9X
 
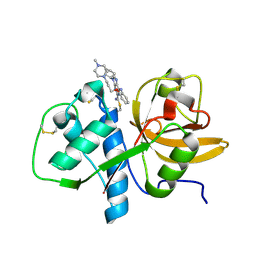 | |
8BET
 
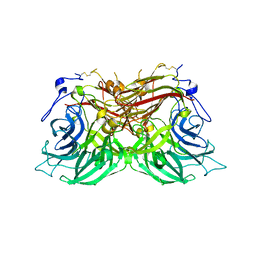 | |
1TX4
 
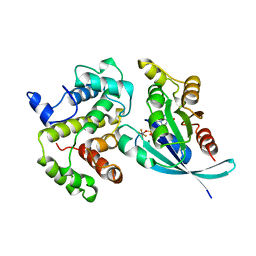 | | RHO/RHOGAP/GDP(DOT)ALF4 COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, P50-RHOGAP, ... | | Authors: | Rittinger, K, Walker, P.A, Smerdon, S.J, Gamblin, S.J. | | Deposit date: | 1997-07-29 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure at 1.65 A of RhoA and its GTPase-activating protein in complex with a transition-state analogue.
Nature, 389, 1997
|
|
8B94
 
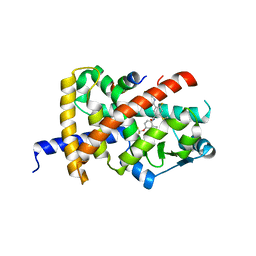 | | Crystal structure of PPARG and NCOR2 with BAY-5516, an inverse agonist | | Descriptor: | Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma, ~{N}3-[4-[bis(fluoranyl)methoxy]-2-methyl-phenyl]-4-chloranyl-6-fluoranyl-~{N}1-[(4-fluorophenyl)methyl]benzene-1,3-dicarboxamide | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Siegel, S, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and characterization of orally bioavailable 4-chloro-6-fluoroisophthalamides as covalent PPARG inverse-agonists.
Bioorg.Med.Chem., 78, 2022
|
|
1UAB
 
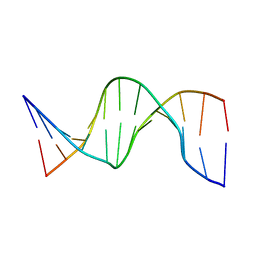 | | NMR structure of hemimethylated GATC site | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*GP*(6MA)P*TP*CP*TP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*TP*CP*TP*GP*CP*G)-3') | | Authors: | Bae, S.-H, Cheong, H.-K, Kang, S, Hwang, D.S, Cheong, C, Choi, B.-S. | | Deposit date: | 2003-03-07 | | Release date: | 2004-04-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of hemimethylated GATC sites: implications for DNA-SeqA recognition
J.Biol.Chem., 278, 2003
|
|
8BFH
 
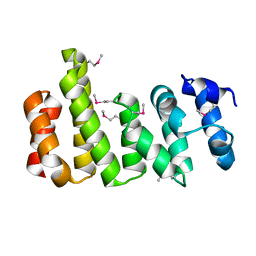 | | CNOT11 | | Descriptor: | CCR4-NOT transcription complex subunit 11 | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The human CNOT1-CNOT10-CNOT11 complex forms a structural platform for protein-protein interactions.
Cell Rep, 42, 2023
|
|
8BE0
 
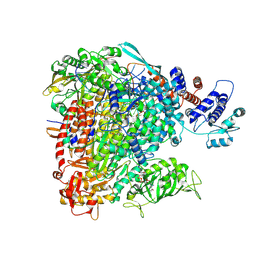 | |
1UAX
 
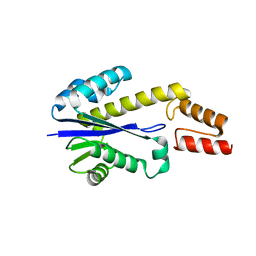 | |
8B2M
 
 | | Matrix-metallopeptidase inhibitor Potempin A (PotA) from Tannerella forsythia | | Descriptor: | NICKEL (II) ION, Tannerella forsythia Potempin A (PotA) | | Authors: | Potempa, J, Ksiazek, M, Goulas, T, Cuppari, A, Rodriguez-Banqueri, A, Arolas, J.L, Lopez-Pelegrin, M, Garcia-Ferrer, I, Guevara, T. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A unique network of attack, defence and competence on the outer membrane of the periodontitis pathogen Tannerella forsythia.
Chem Sci, 14, 2023
|
|
1UB9
 
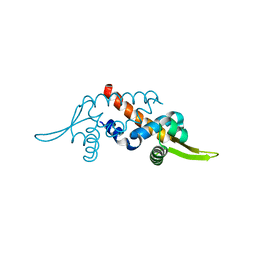 | | Structure of the transcriptional regulator homologue protein from Pyrococcus horikoshii OT3 | | Descriptor: | Hypothetical protein PH1061 | | Authors: | Okada, U, Sakai, N, Tajika, Y, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2003-04-03 | | Release date: | 2004-05-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis of the transcriptional regulator homolog protein from Pyrococcus horikoshii OT3.
Proteins, 63, 2006
|
|
1UGF
 
 | |
