1TJ3
 
 | | X-Ray structure of the Sucrose-Phosphatase (SPP) from Synechocystis sp. PCC6803 in a closed conformation | | Descriptor: | MAGNESIUM ION, Sucrose-Phosphatase | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.-L. | | Deposit date: | 2004-06-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell.
Plant Cell, 17, 2005
|
|
1TJ4
 
 | | X-Ray structure of the Sucrose-Phosphatase (SPP) from Synechocystis sp. PCC6803 in complex with sucrose | | Descriptor: | MAGNESIUM ION, Sucrose-Phosphatase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.-L. | | Deposit date: | 2004-06-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell.
Plant Cell, 17, 2005
|
|
6Y2K
 
 | | Crystal structure of beta-galactosidase from the psychrophilic Marinomonas ef1 | | Descriptor: | CHLORIDE ION, GLYCEROL, beta-galactosidase | | Authors: | Mangiagalli, M, Lapi, M, Maione, S, Orlando, M, Brocca, S, Pesce, A, Barbiroli, A, Pucciarelli, S, Camilloni, C, Lotti, M. | | Deposit date: | 2020-02-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The co-existence of cold activity and thermal stability in an Antarctic GH42 beta-galactosidase relies on its hexameric quaternary arrangement.
Febs J., 288, 2021
|
|
8PKH
 
 | | Borrelia bacteriophage BB1 procapsid, fivefold-symmetrized outer shell | | Descriptor: | Decorator protein P03, Decorator protein P04, Decorator protein P05, ... | | Authors: | Rumnieks, J, Fuzik, T, Tars, K. | | Deposit date: | 2023-06-26 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of the Borrelia Bacteriophage phi BB1 Procapsid.
J.Mol.Biol., 435, 2023
|
|
8A41
 
 | | Nudaurelia capensis omega virus procapsid at pH7.6 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-10 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.88 Å) | | Cite: | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs)
To be published
|
|
3EXU
 
 | | A glycoside hydrolase family 11 xylanase with an extended thumb region | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase, GLYCEROL | | Authors: | Vandermarliere, E, Pollet, A, Strelkov, S.V, Delcour, J.A, Courtin, C.M. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystallographic and activity-based evidence for thumb flexibility and its relevance in glycoside hydrolase family 11 xylanases
Proteins, 77, 2009
|
|
8A3C
 
 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs)
To be published
|
|
2LAV
 
 | | NMR solution structure of human Vaccinia-Related Kinase 1 | | Descriptor: | Vaccinia-related kinase 1 | | Authors: | Shin, J, Yoon, H.S. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Human Vaccinia-related Kinase 1 (VRK1) Reveals the C-terminal Tail Essential for Its Structural Stability and Autocatalytic Activity.
J.Biol.Chem., 286, 2011
|
|
4J38
 
 | | Structure of Borrelia burgdorferi Outer surface protein E in complex with Factor H domains 19-20 | | Descriptor: | Complement factor H, Outer surface protein E, SULFATE ION | | Authors: | Bhattacharjee, A, Kolodziejczyk, R, Kajander, T, Goldman, A, Jokiranta, T.S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structural Basis for Complement Evasion by Lyme Disease Pathogen Borrelia burgdorferi
J.Biol.Chem., 288, 2013
|
|
4CBE
 
 | | Crystal structure of complement factors H and FHL-1 binding protein BBH06 or CRASP-2 from Borrelia burgdorferi (Native) | | Descriptor: | COMPLEMENT REGULATOR-ACQUIRING SURFACE PROTEIN 2 (CRASP-2) | | Authors: | Brangulis, K, Petrovskis, I, Kazaks, A, Ranka, R, Tars, K. | | Deposit date: | 2013-10-13 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Characterization of Cspz, a Complement Regulator Factor H and Fhl-1 Binding Protein from Borrelia Burgdorferi.
FEBS J., 281, 2014
|
|
2LKS
 
 | | Ff11-60 | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Barette, J, Velyvis, A, Religa, T.L, Korzhnev, D.M, Kay, L.E. | | Deposit date: | 2011-10-19 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cross-Validation of the Structure of a Transiently Formed and Low Populated FF Domain Folding Intermediate Determined by Relaxation Dispersion NMR and CS-Rosetta.
J.Phys.Chem.B, 116, 2012
|
|
2L9V
 
 | | NMR structure of the FF domain L24A mutant's folding transition state | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Korzhnev, D.M, Vernon, R.M, Religa, T.L, Hansen, A, Baker, D, Fersht, A.R, Kay, L.E. | | Deposit date: | 2011-02-24 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nonnative interactions in the FF domain folding pathway from an atomic resolution structure of a sparsely populated intermediate: an NMR relaxation dispersion study.
J.Am.Chem.Soc., 133, 2011
|
|
6KU3
 
 | | Crystal structure of gibberellin 2-oxidase3 (GA2ox3)in rice | | Descriptor: | 2-OXOGLUTARIC ACID, GIBBERELLIN A4, GLYCEROL, ... | | Authors: | Takehara, S, Mikami, B, Sakuraba, S, Matsuoka, M, Ueguchi-Tanaka, M. | | Deposit date: | 2019-08-30 | | Release date: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A common allosteric mechanism regulates homeostatic inactivation of auxin and gibberellin.
Nat Commun, 11, 2020
|
|
1W33
 
 | | BbCRASP-1 from Borrelia Burgdorferi | | Descriptor: | BBCRASP-1, GLYCEROL | | Authors: | Cordes, F.S, Roversi, P, Goodstadt, L, Ponting, C, Kraiczy, P, Skerka, C, Kirschfink, M, Simon, M.M, Brade, V, Zipfel, P, Wallich, R, Lea, S.M. | | Deposit date: | 2004-07-13 | | Release date: | 2005-02-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Novel Fold for the Factor H-Binding Protein Bbcrasp-1 of Borrelia Burgdorferi
Nat.Struct.Mol.Biol., 12, 2005
|
|
1W3Z
 
 | | SeMet derivative of BbCRASP-1 from Borrelia Burgdorferi | | Descriptor: | BBCRASP-1 | | Authors: | Cordes, F.S, Roversi, P, Goodstadt, L, Ponting, C, Kraiczy, P, Skerka, C, Kirschfink, M, Simon, M.M, Brade, V, Zipfel, P, Wallich, R, Lea, S.M. | | Deposit date: | 2004-07-21 | | Release date: | 2005-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Novel Fold for the Factor H-Binding Protein Bbcrasp-1 of Borrelia Burgdorferi
Nat.Struct.Mol.Biol., 12, 2005
|
|
8TBE
 
 | | Co-crystal structure of SARS-CoV-2 Mpro with Pomotrelvir | | Descriptor: | 3C-like proteinase nsp5, Pomotrelvir bound form | | Authors: | Olland, A, Fontano, E, White, A. | | Deposit date: | 2023-06-28 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Evaluation of in vitro antiviral activity of SARS-CoV-2 M pro inhibitor pomotrelvir and cross-resistance to nirmatrelvir resistance substitutions.
Antimicrob.Agents Chemother., 67, 2023
|
|
2KC9
 
 | |
6CIP
 
 | |
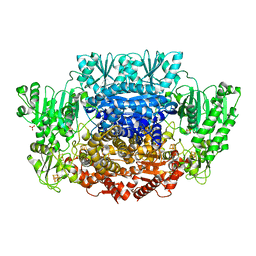
6CIN
 
 | |
6CIQ
 
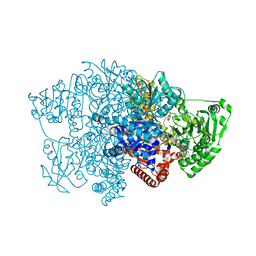 | |
6CIO
 
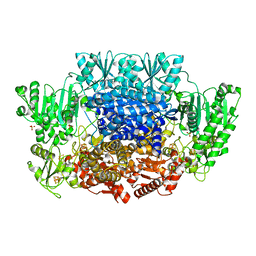 | | Pyruvate:ferredoxin oxidoreductase from Moorella thermoacetica with lactyl-TPP bound | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Chen, P.Y.-T, Drennan, C.L. | | Deposit date: | 2018-02-24 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Binding site for coenzyme A revealed in the structure of pyruvate:ferredoxin oxidoreductase fromMoorella thermoacetica.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2KC8
 
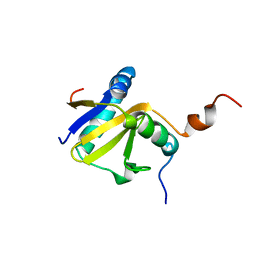 | | Structure of E. coli toxin RelE (R81A/R83A) mutant in complex with antitoxin RelBc (K47-L79) peptide | | Descriptor: | Antitoxin RelB, Toxin relE | | Authors: | Li, G, Zhang, Y, Inouye, M, Ikura, M. | | Deposit date: | 2008-12-17 | | Release date: | 2009-03-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Inhibitory mechanism of Escherichia coli RelE-RelB toxin-antitoxin module involves a helix displacement near an mRNA interferase active site.
J.Biol.Chem., 284, 2009
|
|
5I6L
 
 | | Crystal Structure of Copper Nitrite Reductase at 100K after 2.76 MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Hough, M.A, Strange, R.W. | | Deposit date: | 2016-02-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Serial crystallography captures enzyme catalysis in copper nitrite reductase at atomic resolution from one crystal.
Iucrj, 3, 2016
|
|
5I6K
 
 | |
5I6N
 
 | | Crystal Structure of Copper Nitrite Reductase at 100K after 11.73 MGy | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRIC OXIDE, ... | | Authors: | Horrell, S, Hough, M.A, Strange, R.W. | | Deposit date: | 2016-02-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Serial crystallography captures enzyme catalysis in copper nitrite reductase at atomic resolution from one crystal.
Iucrj, 3, 2016
|
|
