7S8B
 
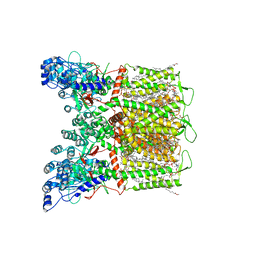 | | Cryo-EM structure of human TRPV6 in complex with channel blocker ruthenium red | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2021-09-17 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Structural mechanisms of TRPV6 inhibition by ruthenium red and econazole.
Nat Commun, 12, 2021
|
|
2J1Y
 
 | |
2J5C
 
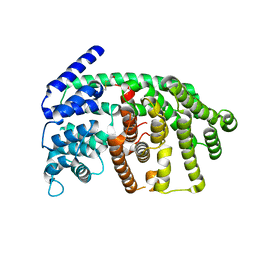 | | Rational conversion of substrate and product specificity in a monoterpene synthase. Structural insights into the molecular basis of rapid evolution. | | Descriptor: | 1,8-CINEOLE SYNTHASE, BETA-MERCAPTOETHANOL | | Authors: | Kampranis, S.C, Ioannidis, D, Purvis, A, Mahrez, W, Ninga, E, Katerelos, N.A, Anssour, S, Dunwell, J.M, Makris, A.M, Goodenough, P.W, Johnson, C.B. | | Deposit date: | 2006-09-14 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational Conversion of Substrate and Product Specificity in a Salvia Monoterpene Synthase: Structural Insights Into the Evolution of Terpene Synthase Function.
Plant Cell, 19, 2007
|
|
4DCG
 
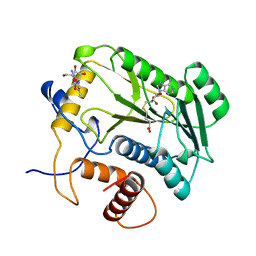 | | VACCINIA METHYLTRANSFERASE VP39 MUTANT D182A COMPLEXED WITH M7G AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 7-METHYLGUANOSINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1998-07-13 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
7SN3
 
 | | Structure of human SARS-CoV-2 spike glycoprotein trimer bound by neutralizing antibody C1C-A3 Fab (variable region) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Abraham, J, Shankar, S. | | Deposit date: | 2021-10-27 | | Release date: | 2021-12-08 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
4DBV
 
 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH LEU 33 REPLACED BY THR, THR 34 REPLACED BY GLY, ASP 36 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NADP+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1997-01-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
4DAA
 
 | |
3LXV
 
 | | Tyrosine 447 of Protocatechuate 3,4-Dioxygenase Controls Efficient Progress Through Catalysis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-NITROCATECHOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Purpero, V.M, Lipscomb, J.D. | | Deposit date: | 2010-02-25 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tyrosine 447 of Protocatechuate 3,4-Dioxygenase Controls Efficient Progress Through Catalysis
To be Published, 2010
|
|
2IWT
 
 | | Thioredoxin h2 (HvTrxh2) in a mixed disulfide complex with the target protein BASI | | Descriptor: | ALPHA-AMYLASE/SUBTILISIN INHIBITOR, CITRATE ANION, THIOREDOXIN H ISOFORM 2 | | Authors: | Maeda, K, Hagglund, P, Finnie, C, Svensson, B, Henriksen, A. | | Deposit date: | 2006-07-04 | | Release date: | 2006-11-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Target Protein Recognition by the Protein Disulfide Reductase Thioredoxin.
Structure, 14, 2006
|
|
2J20
 
 | |
2J6T
 
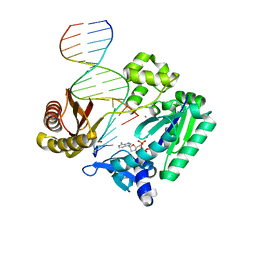 | | Ternary complex of Sulfolobus solfataricus Dpo4 DNA polymerase, O6- methylguanine modified DNA, and dATP. | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*C)-3', 5'-D(*TP*CP*AP*TP*XP*GP*AP*AP*TP*CP *CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Eoff, R.L, Guengerich, F.P, Egli, M. | | Deposit date: | 2006-10-04 | | Release date: | 2006-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sulfolobus Solfataricus DNA Polymerase Dpo4 is Partially Inhibited by "Wobble" Pairing between O6- Methylguanine and Cytosine, But Accurate Bypass is Preferred.
J.Biol.Chem., 282, 2007
|
|
7S6Q
 
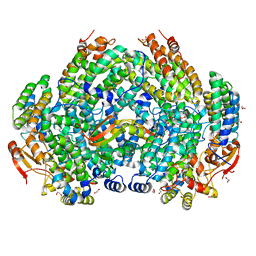 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit DBL2 | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, FE (III) ION, ... | | Authors: | Johns, J.C, Banerjee, R, Semonis, M.M, Shi, K, Aihara, H, Lipscomb, J.D. | | Deposit date: | 2021-09-14 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray Crystal Structures of Methane Monooxygenase Hydroxylase Complexes with Variants of Its Regulatory Component: Correlations with Altered Reaction Cycle Dynamics.
Biochemistry, 61, 2022
|
|
3LVA
 
 | |
4E7D
 
 | |
4E85
 
 | |
7S6T
 
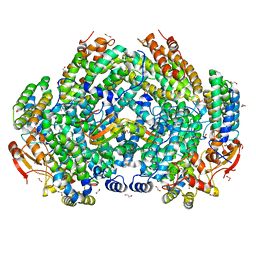 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit H33A | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, FE (III) ION, ... | | Authors: | Johns, J.C, Banerjee, R, Semonis, M.M, Shi, K, Aihara, H, Lipscomb, J.D. | | Deposit date: | 2021-09-14 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | X-ray Crystal Structures of Methane Monooxygenase Hydroxylase Complexes with Variants of Its Regulatory Component: Correlations with Altered Reaction Cycle Dynamics.
Biochemistry, 61, 2022
|
|
2J4X
 
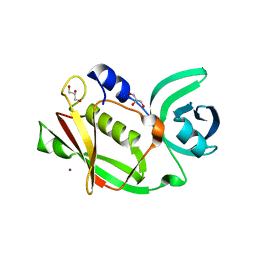 | | Streptococcus dysgalactiae-derived mitogen (SDM) | | Descriptor: | GLYCEROL, MITOGEN, ZINC ION | | Authors: | Saarinen, S, Kato, H, Uchiyama, T, Miyoshi-Akiyama, T, Papageorgiou, A.C. | | Deposit date: | 2006-09-07 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Streptococcus Dysgalactiae-Derived Mitogen Reveals a Zinc-Binding Site and Alterations in Tcr Binding.
J.Mol.Biol., 373, 2007
|
|
7S6R
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit with H5A mutation | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, FE (III) ION, ... | | Authors: | Johns, J.C, Banerjee, R, Semonis, M.M, Shi, K, Aihara, H, Lipscomb, J.D. | | Deposit date: | 2021-09-14 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | X-ray Crystal Structures of Methane Monooxygenase Hydroxylase Complexes with Variants of Its Regulatory Component: Correlations with Altered Reaction Cycle Dynamics.
Biochemistry, 61, 2022
|
|
7S6S
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit DBL1 | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, FE (III) ION, ... | | Authors: | Johns, J.C, Banerjee, R, Semonis, M.M, Shi, K, Aihara, H, Lipscomb, J.D. | | Deposit date: | 2021-09-14 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray Crystal Structures of Methane Monooxygenase Hydroxylase Complexes with Variants of Its Regulatory Component: Correlations with Altered Reaction Cycle Dynamics.
Biochemistry, 61, 2022
|
|
2J6S
 
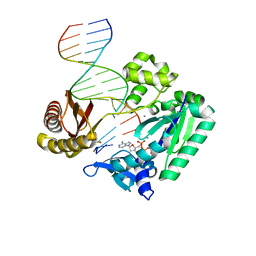 | | Ternary complex of Sulfolobus solfataricus Dpo4 DNA polymerase, O6- methylguanine modified DNA, and dATP. | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*C)-3', 5'-D(*TP*CP*AP*TP*XP*GP*AP*AP*TP*CP *CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Eoff, R.L, Guengerich, F.P, Egli, M. | | Deposit date: | 2006-10-04 | | Release date: | 2006-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sulfolobus Solfataricus DNA Polymerase Dpo4 is Partially Inhibited by "Wobble" Pairing between O6- Methylguanine and Cytosine, But Accurate Bypass is Preferred.
J.Biol.Chem., 282, 2007
|
|
7S63
 
 | |
2J5K
 
 | | 2.0 A resolution structure of the wild type malate dehydrogenase from Haloarcula marismortui (radiation damage series) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE | | Authors: | Fioravanti, E, Vellieux, F.M.D, Amara, P, Madern, D, Weik, M. | | Deposit date: | 2006-09-18 | | Release date: | 2006-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specific Radiation Damage to Acidic Residues and its Relation to Their Chemical and Structural Environment.
J.Synchrotron Radiat., 14, 2007
|
|
3LW5
 
 | | Improved model of plant photosystem I | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, AT3g54890, BETA-CAROTENE, ... | | Authors: | Nelson, N, Toporik, H. | | Deposit date: | 2010-02-23 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure determination and improved model of plant photosystem I
J.Biol.Chem., 285, 2010
|
|
4MEH
 
 | |
2J4R
 
 | | Structural Study of the Aquifex aeolicus PPX-GPPA enzyme | | Descriptor: | EXOPOLYPHOSPHATASE, GUANOSINE-5',3'-TETRAPHOSPHATE | | Authors: | Kristensen, O. | | Deposit date: | 2006-09-05 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of the Ppx/Gppa Phosphatase from Aquifex Aeolicus in Complex with the Alarmone Ppgpp
J.Mol.Biol., 375, 2008
|
|
