8P3A
 
 | |
5EWX
 
 | | Fusion protein of T4 lysozyme and B4 domain of protein A from staphylococcal aureus with chemical cross-linker EY-CBS | | Descriptor: | 2,2'-ethyne-1,2-diylbis{5-[(chloroacetyl)amino]benzenesulfonic acid}, Endolysin,Immunoglobulin G-binding protein A,Endolysin | | Authors: | Jeong, W.H, Lee, H, Song, D.H, Lee, J.O. | | Deposit date: | 2015-11-22 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Connecting two proteins using a fusion alpha helix stabilized by a chemical cross linker.
Nat Commun, 7, 2016
|
|
5XSZ
 
 | | Crystal structure of zebrafish lysophosphatidic acid receptor LPA6 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lysophosphatidic acid receptor 6a,Endolysin,Lysophosphatidic acid receptor 6a | | Authors: | Taniguchi, R, Nishizawa, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-16 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into ligand recognition by the lysophosphatidic acid receptor LPA6
Nature, 548, 2017
|
|
4TN3
 
 | | Structure of the BBox-Coiled-coil region of Rhesus Trim5alpha | | Descriptor: | TRIM5/cyclophilin A fusion protein/T4 Lysozyme chimera, ZINC ION | | Authors: | Kirkpatrick, J.J, Stoye, J.P, Taylor, I.A, Goldstone, D.C. | | Deposit date: | 2014-06-03 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1989 Å) | | Cite: | Structural studies of postentry restriction factors reveal antiparallel dimers that enable avid binding to the HIV-1 capsid lattice.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8EIT
 
 | | Structure of FFAR1-Gq complex bound to DHA | | Descriptor: | A modified Guanine nucleotide-binding protein G(q) subunit alpha, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 1, ... | | Authors: | Kumari, P, Inoue, A, Chapman, K, Lian, P, Rosenbaum, D.M. | | Deposit date: | 2022-09-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular mechanism of fatty acid activation of FFAR1.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7L3J
 
 | | T4 Lysozyme L99A - benzylacetate - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3A
 
 | | T4 Lysozyme L99A - toluene - cryo | | Descriptor: | Endolysin, TOLUENE | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L39
 
 | | T4 Lysozyme L99A - toluene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L38
 
 | | T4 Lysozyme L99A - Apo - cryo | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3B
 
 | | T4 Lysozyme L99A - iodobenzene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L37
 
 | | T4 Lysozyme L99A - Apo - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.439 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3G
 
 | | T4 Lysozyme L99A - 4-iodotoluene - cryo | | Descriptor: | 1-iodo-4-methylbenzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3F
 
 | | T4 Lysozyme L99A - 4-iodotoluene - RT | | Descriptor: | 1-iodo-4-methylbenzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3H
 
 | | T4 Lysozyme L99A - ethylbenzene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3E
 
 | | T4 Lysozyme L99A - 3-iodotoluene - cryo | | Descriptor: | 1-iodo-3-methylbenzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3K
 
 | | T4 Lysozyme L99A - benzylacetate - cryo | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3C
 
 | | T4 Lysozyme L99A - o-xylene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3D
 
 | | T4 Lysozyme L99A - 3-iodotoluene - RT | | Descriptor: | 1-iodo-3-methylbenzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3I
 
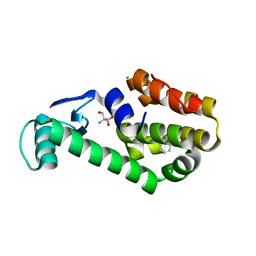 | | T4 Lysozyme L99A - propylbenzene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Endolysin, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7XB5
 
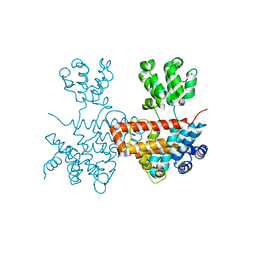 | |
8A5X
 
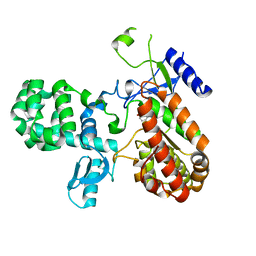 | |
7Z36
 
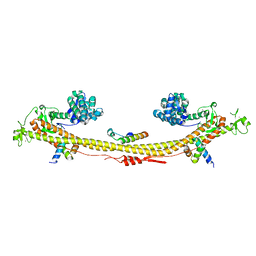 | | Crystal structure of the KAP1 tripartite motif in complex with the ZNF93 KRAB domain | | Descriptor: | Endolysin,Transcription intermediary factor 1-beta,Isoform 2 of Transcription intermediary factor 1-beta, SMARCAD1 CUE1 domain, ZINC ION, ... | | Authors: | Stoll, G.A, Modis, Y. | | Deposit date: | 2022-03-01 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and functional mapping of the KRAB-KAP1 repressor complex.
Embo J., 41, 2022
|
|
2A1L
 
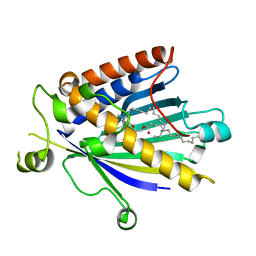 | | Rat PITP-Beta Complexed to Phosphatidylcholine | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Phosphatidylinositol transfer protein beta isoform | | Authors: | Vordtriede, P.B, Doan, C.N, Tremblay, J.M, Helmkamp, G.M, Yoder, M.D. | | Deposit date: | 2005-06-20 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure of PITPbeta in Complex with Phosphatidylcholine: Comparison of Structure and Lipid Transfer to Other PITP Isoforms.
Biochemistry, 44, 2005
|
|
2AA2
 
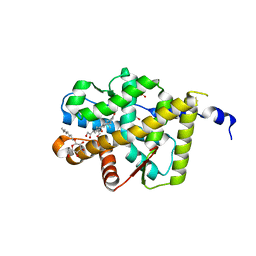 | | Mineralocorticoid Receptor with Bound Aldosterone | | Descriptor: | ALDOSTERONE, GLYCEROL, Mineralocorticoid receptor, ... | | Authors: | Bledsoe, R.K, Madauss, K.P, Holt, J.A, Apolito, C.J, Lambert, M.H, Pearce, K.H, Stanley, T.B, Stewart, E.L, Trump, R.P, Willson, T.M, Williams, S.P. | | Deposit date: | 2005-07-13 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Ligand-mediated Hydrogen Bond Network Required for the Activation of the Mineralocorticoid Receptor
J.Biol.Chem., 280, 2005
|
|
2OBD
 
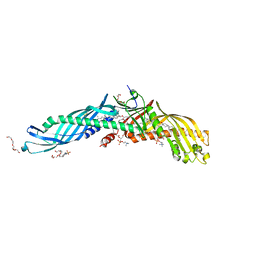 | | Crystal Structure of Cholesteryl Ester Transfer Protein | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Qiu, X. | | Deposit date: | 2006-12-18 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules.
Nat.Struct.Mol.Biol., 14, 2007
|
|
