1N2Z
 
 | | 2.0 Angstrom structure of BtuF, the vitamin B12 binding protein of E. coli | | Descriptor: | CADMIUM ION, CHLORIDE ION, CYANOCOBALAMIN, ... | | Authors: | Borths, E.L, Locher, K.P, Lee, A.T, Rees, D.C. | | Deposit date: | 2002-10-24 | | Release date: | 2002-12-18 | | Last modified: | 2021-08-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of Escherichia coli BtuF and binding to its cognate ATP binding cassette transporter
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JZF
 
 | | Pseudomonas aeruginosa Oxidized Azurin(Cu2+) Ru(tpy)(phen)(His83) | | Descriptor: | (2,2':6',2'-TERPYRIDINE)-(1,10-PHENANTHROLINE) RUTHENIUM (II), AZURIN, COPPER (II) ION, ... | | Authors: | Crane, B.R, Di Bilio, A.J, Winkler, J.R, Gray, H.B. | | Deposit date: | 2001-09-16 | | Release date: | 2001-10-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Electron tunneling in single crystals of Pseudomonas aeruginosa azurins.
J.Am.Chem.Soc., 123, 2001
|
|
1JZI
 
 | | Pseudomonas aeruginosa Azurin Re(phen)(CO)3(His83) | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), AZURIN, COPPER (II) ION, ... | | Authors: | Crane, B.R, Di Bilio, A.J, Winkler, J.R, Gray, H.B. | | Deposit date: | 2001-09-16 | | Release date: | 2001-10-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Electron tunneling in single crystals of Pseudomonas aeruginosa azurins.
J.Am.Chem.Soc., 123, 2001
|
|
2BO0
 
 | | Crystal structure of the C130A mutant of nitrite reductase from Alcaligenes xylosoxidans | | Descriptor: | DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, TETRAETHYLENE GLYCOL, ZINC ION | | Authors: | Hough, M.A, Ellis, M.J, Antonyuk, S, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-04-06 | | Release date: | 2006-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High Resolution Structural Studies of Mutants Provide Insights Into Catalysis and Electron Transfer Processes in Copper Nitrite Reductase
J.Mol.Biol., 350, 2005
|
|
1LPI
 
 | | HEW LYSOZYME: TRP...NA CATION-PI INTERACTION | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Wouters, J. | | Deposit date: | 1998-04-15 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cation-pi (Na+-Trp) interactions in the crystal structure of tetragonal lysozyme.
Protein Sci., 7, 1998
|
|
2H92
 
 | |
1PM5
 
 | | Crystal structure of wild type Lactococcus lactis Fpg complexed to a tetrahydrofuran containing DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*TP*TP*(3DR)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Pereira de Jesus-Tran, K, Serre, L, Zelwer, C, Castaing, B. | | Deposit date: | 2003-06-10 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA.
Nucleic Acids Res., 33, 2005
|
|
1V0Y
 
 | |
1LCN
 
 | | Monoclinic hen egg white lysozyme, thiocyanate complex | | Descriptor: | PROTEIN (LYSOZYME), THIOCYANATE ION | | Authors: | Hamiaux, C, Prange, T, Ducruix, A, Vaney, M.C. | | Deposit date: | 1998-10-27 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural effects of monovalent anions on polymorphic lysozyme crystals.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2HSB
 
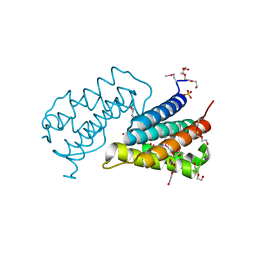 | |
2HKD
 
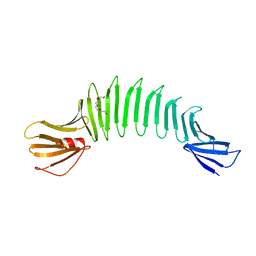 | |
2HL7
 
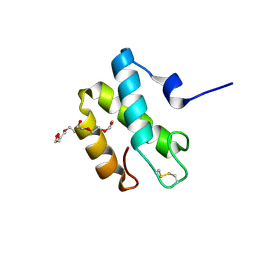 | | Crystal structure of the periplasmic domain of CcmH from Pseudomonas aeruginosa | | Descriptor: | Cytochrome C-type biogenesis protein CcmH, TETRAETHYLENE GLYCOL | | Authors: | Di Matteo, A, Travaglini-Allocatelli, C, Gianni, S, Brunori, M. | | Deposit date: | 2006-07-06 | | Release date: | 2007-07-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A strategic protein in cytochrome c maturation: three-dimensional structure of CcmH and binding to apocytochrome c
J.Biol.Chem., 282, 2007
|
|
2CNW
 
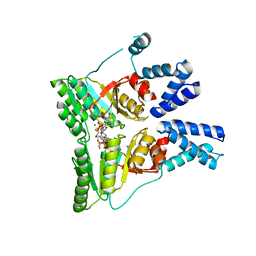 | | GDPALF4 complex of the SRP GTPases Ffh and FtsY | | Descriptor: | CELL DIVISION PROTEIN FTSY, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Focia, P.J, Gawronski-Salerno, J, Coon V, J.S, Freymann, D.M. | | Deposit date: | 2006-05-24 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of a Gdp:Alf(4) Complex of the Srp Gtpases Ffh and Ftsy, and Identification of a Peripheral Nucleotide Interaction Site.
J.Mol.Biol., 360, 2006
|
|
2CM6
 
 | | crystal structure of the C2B domain of rabphilin3A | | Descriptor: | CALCIUM ION, PHOSPHATE ION, RABPHILIN-3A | | Authors: | Schlicker, C, Montaville, P, Sheldrick, G.M, Becker, S. | | Deposit date: | 2006-05-04 | | Release date: | 2006-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The C2A-C2B Linker Defines the High Affinity Ca2+ Binding Mode of Rabphilin-3A.
J.Biol.Chem., 282, 2007
|
|
2CAQ
 
 | | Structure of R21L mutant of Sh28GST in complex with GSH | | Descriptor: | BETA-MERCAPTOETHANOL, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE 28 KDA, ... | | Authors: | Baiocco, P, Gourlay, L.J, Angelucci, F, Bellelli, A, Miele, A.E, Brunori, M. | | Deposit date: | 2005-12-22 | | Release date: | 2006-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the Mechanism of Gsh Activation in Schistosoma Haematobium Glutathione-S-Transferase by Site-Directed Mutagenesis and X-Ray Crystallography.
J.Mol.Biol., 360, 2006
|
|
1TBW
 
 | | Ligand Induced Conformational Shift in the N-terminal Domain of GRP94, Open Conformation | | Descriptor: | ADENOSINE MONOPHOSPHATE, Endoplasmin, MAGNESIUM ION, ... | | Authors: | Gewirth, D.T, Immormino, R.M, Dollins, D.E, Shaffer, P.L, Walker, M.A, Soldano, K.L. | | Deposit date: | 2004-05-20 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ligand-induced Conformational Shift in the N-terminal Domain of GRP94, an Hsp90 Chaperone.
J.Biol.Chem., 279, 2004
|
|
1Q69
 
 | | Solution structure of T-cell surface glycoprotein CD8 alpha chain and Proto-oncogene tyrosine-protein kinase LCK fragments | | Descriptor: | Proto-oncogene tyrosine-protein kinase LCK, T-cell surface glycoprotein CD8 alpha chain, ZINC ION | | Authors: | Kim, P.W, Sun, Z.Y, Blacklow, S.C, Wagner, G, Eck, M.J. | | Deposit date: | 2003-08-12 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A zinc clasp structure tethers Lck to T cell coreceptors CD4 and CD8.
Science, 301, 2003
|
|
1L5Y
 
 | | CRYSTAL STRUCTURE OF MG2+ / BEF3-BOUND RECEIVER DOMAIN OF SINORHIZOBIUM MELILOTI DCTD | | Descriptor: | BERYLLIUM DIFLUORIDE, BERYLLIUM TETRAFLUORIDE ION, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Park, S, Meyer, M, Jones, A.D, Yennawar, H.P, Yennawar, N.H, Nixon, B.T. | | Deposit date: | 2002-03-08 | | Release date: | 2002-10-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two-component signaling in the AAA + ATPase DctD: binding Mg2+ and BeF3- selects between alternate dimeric states of the receiver domain
FASEB J., 16, 2002
|
|
2I7D
 
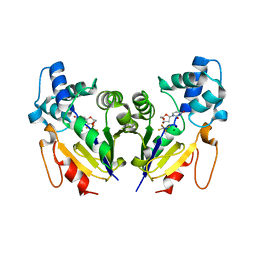 | |
1SW4
 
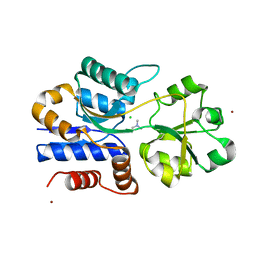 | | Crystal structure of ProX from Archeoglobus fulgidus in complex with trimethyl ammonium | | Descriptor: | CHLORIDE ION, TETRAMETHYLAMMONIUM ION, ZINC ION, ... | | Authors: | Schiefner, A, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2004-03-30 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the binding of compatible solutes by ProX from the hyperthermophilic archaeon Archaeoglobus fulgidus.
J.Biol.Chem., 279, 2004
|
|
2I5Z
 
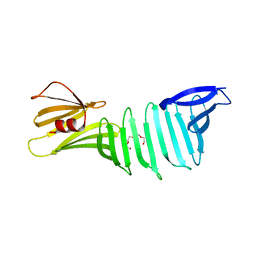 | | The crystal structure of OspA mutant | | Descriptor: | Outer surface protein A, TETRAETHYLENE GLYCOL | | Authors: | Makabe, K, Terechko, V, Koide, S. | | Deposit date: | 2006-08-26 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Hydrophobic surface burial is the major stability determinant of a flat, single-layer beta-sheet.
J.Mol.Biol., 368, 2007
|
|
2F90
 
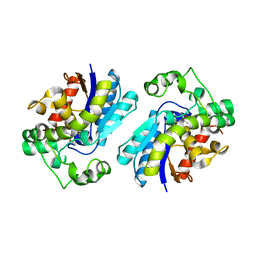 | | Crystal structure of bisphosphoglycerate mutase in complex with 3-phosphoglycerate and AlF4- | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Bisphosphoglycerate mutase, TETRAFLUOROALUMINATE ION | | Authors: | Wang, Y, Liu, L, Wei, Z, Gong, W. | | Deposit date: | 2005-12-05 | | Release date: | 2006-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Seeing the process of histidine phosphorylation in human bisphosphoglycerate mutase
J.Biol.Chem., 281, 2006
|
|
1OE1
 
 | |
1OE3
 
 | | Atomic resolution structure of 'Half Apo' NiR | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, TETRAETHYLENE GLYCOL | | Authors: | Ellis, M.J, Dodd, F.E, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2003-03-18 | | Release date: | 2004-07-21 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic Resolution Structures of Native Copper Nitrite Reductase from Alcaligenes Xylosoxidans and the Active Site Mutant Asp92Glu
J.Mol.Biol., 328, 2003
|
|
1UN8
 
 | | Crystal structure of the dihydroxyacetone kinase of C. freundii (native form) | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, DIHYDROXYACETONE KINASE | | Authors: | Siebold, C, Arnold, I, Garcia-Alles, L.F, Baumann, U, Erni, B. | | Deposit date: | 2003-09-08 | | Release date: | 2003-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Citrobacter Freundii Dihydroxyacetone Kinase Reveals an Eight-Stranded Alpha-Helical Barrel ATP-Binding Domain
J.Biol.Chem., 278, 2003
|
|
