4XEE
 
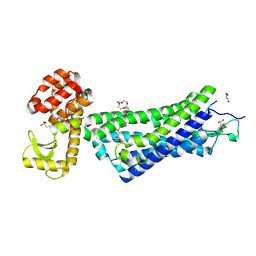 | | Structure of active-like neurotensin receptor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krumm, B.E, White, J.F, Shah, P, Grisshammer, R. | | Deposit date: | 2014-12-23 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural prerequisites for G-protein activation by the neurotensin receptor.
Nat Commun, 6, 2015
|
|
4YX7
 
 | |
6KK1
 
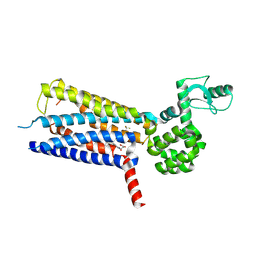 | | Structure of thermal-stabilised(M8) human GLP-1 receptor transmembrane domain | | Descriptor: | Glucagon-like peptide 1 receptor,Endolysin,Glucagon-like peptide 1 receptor, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine | | Authors: | Song, G. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutagenesis facilitated crystallization of GLP-1R.
Iucrj, 6, 2019
|
|
4YXC
 
 | |
4XES
 
 | | Structure of active-like neurotensin receptor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krumm, B.E, White, J.F, Shah, P, Grisshammer, R. | | Deposit date: | 2014-12-24 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural prerequisites for G-protein activation by the neurotensin receptor.
Nat Commun, 6, 2015
|
|
4YXA
 
 | |
6KK7
 
 | | Structure of thermal-stabilised(M6) human GLP-1 receptor transmembrane domain | | Descriptor: | Glucagon-like peptide 1 receptor,Endolysin,Glucagon-like peptide 1 receptor, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine | | Authors: | Song, G. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mutagenesis facilitated crystallization of GLP-1R.
Iucrj, 6, 2019
|
|
5G27
 
 | | Structure of Spin-labelled T4 lysozyme mutant L118C-R1 at Room Temperature | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, ENDOLYSIN, ... | | Authors: | Gohlke, U, Consentius, P, Loll, B, Mueller, R, Kaupp, M, Heinemann, U, Risse, T. | | Deposit date: | 2016-04-07 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Tracking Transient Conformational States of T4 Lysozyme at Room Temperature Combining X-Ray Crystallography and Site-Directed Spin Labeling.
J.Am.Chem.Soc., 138, 2016
|
|
6KJV
 
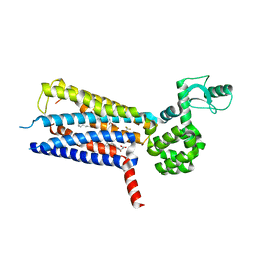 | | Structure of thermal-stabilised(M9) human GLP-1 receptor transmembrane domain | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Glucagon-like peptide 1 receptor,Endolysin,Glucagon-like peptide 1 receptor, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine | | Authors: | Song, G. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutagenesis facilitated crystallization of GLP-1R.
Iucrj, 6, 2019
|
|
5JGZ
 
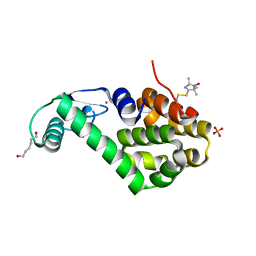 | | Spin-Labeled T4 Lysozyme Construct T151V1 | | Descriptor: | CHLORIDE ION, Endolysin, HEXANE-1,6-DIOL, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
4IAP
 
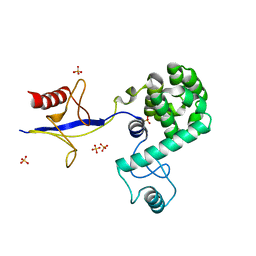 | | Crystal structure of PH domain of Osh3 from Saccharomyces cerevisiae | | Descriptor: | Oxysterol-binding protein homolog 3,Endolysin,Oxysterol-binding protein homolog 3, SULFATE ION | | Authors: | Tong, J, Im, Y.J. | | Deposit date: | 2012-12-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of osh3 reveals a conserved mode of phosphoinositide binding in oxysterol-binding proteins
Structure, 21, 2013
|
|
5JWT
 
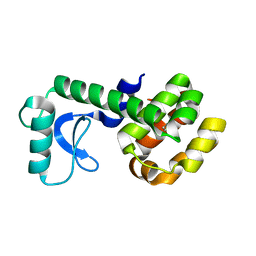 | | T4 Lysozyme L99A/M102Q with Benzene Bound | | Descriptor: | BENZENE, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5JWU
 
 | | T4 Lysozyme L99A/M102Q with 1,2-Dihydro-1,2-azaborine Bound | | Descriptor: | 1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5JWS
 
 | | T4 Lysozyme L99A with 1-Hydro-2-ethyl-1,2-azaborine Bound | | Descriptor: | 2-ethyl-1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5KHZ
 
 | | PSEUDO T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Endolysin | | Authors: | Scholfield, M.R. | | Deposit date: | 2016-06-16 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure-Energy Relationships of Halogen Bonds in Proteins.
Biochemistry, 56, 2017
|
|
5KI8
 
 | | PSEUDO T4 LYSOZYME MUTANT - Y88PHE-BR | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Endolysin | | Authors: | Scholfield, M.R. | | Deposit date: | 2016-06-16 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Energy Relationships of Halogen Bonds in Proteins.
Biochemistry, 56, 2017
|
|
5JWW
 
 | | T4 Lysozyme L99A/M102Q with 1-Hydro-2-ethyl-1,2-azaborine Bound | | Descriptor: | 2-ethyl-1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5KI1
 
 | | PSEUDO T4 LYSOZYME MUTANT - Y18F | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Endolysin | | Authors: | Scholfield, M.R. | | Deposit date: | 2016-06-16 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure-Energy Relationships of Halogen Bonds in Proteins.
Biochemistry, 56, 2017
|
|
5KI2
 
 | |
5KI3
 
 | |
5KGR
 
 | | Spin-Labeled T4 Lysozyme Construct I9V1/V131V1 (30 days) | | Descriptor: | CHLORIDE ION, Endolysin, HEXANE-1,6-DIOL, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-06-13 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.473 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
5KIG
 
 | | PSEUDO T4 LYSOZYME MUTANT - Y88F | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Endolysin | | Authors: | Scholfield, M.R. | | Deposit date: | 2016-06-16 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Energy Relationships of Halogen Bonds in Proteins.
Biochemistry, 56, 2017
|
|
5KII
 
 | | PSEUDO T4 LYSOZYME MUTANT - Y88PHE-METHYL | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Endolysin, PHOSPHATE ION | | Authors: | Scholfield, M.R. | | Deposit date: | 2016-06-16 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure-Energy Relationships of Halogen Bonds in Proteins.
Biochemistry, 56, 2017
|
|
5KIM
 
 | | PSEUDO T4 LYSOZYME MUTANT - Y88PHE-I | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Endolysin, SODIUM ION | | Authors: | Scholfield, M.R. | | Deposit date: | 2016-06-16 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Energy Relationships of Halogen Bonds in Proteins.
Biochemistry, 56, 2017
|
|
5KIO
 
 | | PSEUDO T4 LYSOZYME MUTANT - Y18PHE-I | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Endolysin | | Authors: | Scholfield, M.R. | | Deposit date: | 2016-06-16 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure-Energy Relationships of Halogen Bonds in Proteins.
Biochemistry, 56, 2017
|
|
