5D9T
 
 | |
6ZS1
 
 | | Chaetomium thermophilum CuZn-superoxide dismutase | | Descriptor: | COPPER (II) ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Papageorgiou, A.C, Mohsin, I. | | Deposit date: | 2020-07-15 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal Structure of a Cu,Zn Superoxide Dismutase From the Thermophilic Fungus Chaetomium thermophilum.
Protein Pept.Lett., 28, 2021
|
|
4OWC
 
 | | PtI6 binding to HEWL | | Descriptor: | CHLORIDE ION, IODIDE ION, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Starkey, V.L, Lamplough, L, Kaenket, S, Helliwell, J.R. | | Deposit date: | 2014-01-31 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The binding of platinum hexahalides (Cl, Br and I) to hen egg-white lysozyme and the chemical transformation of the PtI6 octahedral complex to a PtI3 moiety bound to His15.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
5DJF
 
 | |
5DRF
 
 | | Green/cyan WasCFP-pH5.5 at pH 5.5 | | Descriptor: | GLYCEROL, SODIUM ION, WasCFP-pH5.5 at pH 5.5 | | Authors: | Pletnev, V.Z, Pletneva, N.V, Pletnev, S.V. | | Deposit date: | 2015-09-15 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Crystal structure of pH and T dependent green fluorescent protein WasCFP with Trp based chromophore
Russ.J.Bioorganic Chem., 42 (6), 2016
|
|
5DLL
 
 | | Aminopeptidase N (pepN) from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | Aminopeptidase N, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Borek, D, Raczynska, J, Dubrovska, I, Grimshaw, S, Minasov, G, Shuvalova, L, Kwon, K, Anderson, W.F, Otwinowski, Z, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-07 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Aminopeptidase N (pepN) from Francisella tularensis subsp. tularensis SCHU S4
To Be Published
|
|
5DQB
 
 | | Green/cyan WasCFP at pH 8.0 | | Descriptor: | GLYCEROL, SODIUM ION, WasCFP_pH2 | | Authors: | Pletnev, V, Pletneva, N, Pletnev, S. | | Deposit date: | 2015-09-14 | | Release date: | 2016-07-27 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of pH and T dependent green fluorescent protein WasCFP with Trp based chromophore
Russ.J.Bioorganic Chem., 42 (6), 2016
|
|
5DJJ
 
 | | Structure of M. tuberculosis CysQ, a PAP phosphatase with PO4 and 2Mg bound | | Descriptor: | 3'-phosphoadenosine 5'-phosphate phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Fisher, A.J, Erickson, A.I. | | Deposit date: | 2015-09-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal Structures of Mycobacterium tuberculosis CysQ, with Substrate and Products Bound.
Biochemistry, 54, 2015
|
|
5DKT
 
 | | N-terminal His tagged apPOL exonuclease mutant | | Descriptor: | Prex DNA polymerase, SODIUM ION, SULFATE ION | | Authors: | Milton, M.E, Honzatko, R.B, Choe, J.Y, Nelson, S.W. | | Deposit date: | 2015-09-04 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Apicoplast DNA Polymerase from Plasmodium falciparum: The First Look at a Plastidic A-Family DNA Polymerase.
J.Mol.Biol., 428, 2016
|
|
5DLD
 
 | |
4OJ1
 
 | |
5DRH
 
 | | Crystal structure of apo C-As lyase | | Descriptor: | Glyoxalase/bleomycin resistance protein/dioxygenase, SODIUM ION | | Authors: | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2015-09-15 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of apo C-As lyase
To Be Published
|
|
7QRU
 
 | | Structure of Bacillus pseudofirmus Mrp antiporter complex, monomer | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Na(+)/H(+) antiporter subunit B, Na(+)/H(+) antiporter subunit C, ... | | Authors: | Lee, Y. | | Deposit date: | 2022-01-12 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Ion transfer mechanisms in Mrp-type antiporters from high resolution cryoEM and molecular dynamics simulations.
Nat Commun, 13, 2022
|
|
6VSU
 
 | | Arginase from Arabidopsis thaliana in Complex with Ornithine | | Descriptor: | Arginase 1, mitochondrial, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B. | | Deposit date: | 2020-02-11 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Neighboring Subunit Is Engaged to Stabilize the Substrate in the Active Site of Plant Arginases.
Front Plant Sci, 11, 2020
|
|
6D9X
 
 | | Discovery of Potent 2-Aryl-6,7-Dihydro-5HPyrrolo[ 1,2-a]imidazoles as WDR5 WIN-site Inhibitors Using Fragment-Based Methods and Structure-Based Design | | Descriptor: | 2-phenyl-6,7-dihydro-5H-pyrrolo[1,2-a]imidazole, SODIUM ION, WD repeat-containing protein 5 | | Authors: | Phan, J, Fesik, S.W. | | Deposit date: | 2018-04-30 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of Potent 2-Aryl-6,7-dihydro-5 H-pyrrolo[1,2- a]imidazoles as WDR5-WIN-Site Inhibitors Using Fragment-Based Methods and Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
6D55
 
 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | 6-chloro-2-(2,6-diazaspiro[3.3]heptan-2-yl)-1-[(4-fluoro-3,5-dimethylphenyl)methyl]-4-(4-methylpiperazin-1-yl)-1H-benzimidazole, FORMIC ACID, GLYCEROL, ... | | Authors: | Phan, J, Hodges, T, Fesik, S.W. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery and Structure-Based Optimization of Benzimidazole-Derived Activators of SOS1-Mediated Nucleotide Exchange on RAS.
J. Med. Chem., 61, 2018
|
|
2FV7
 
 | | Crystal structure of human ribokinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ribokinase, ... | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-30 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human ribokinase
to be published
|
|
6DAM
 
 | | Crystal structure of lanthanide-dependent methanol dehydrogenase XoxF from Methylomicrobium buryatense 5G | | Descriptor: | LANTHANUM (III) ION, Lanthanide-dependent methanol dehydrogenase XoxF, PYRROLOQUINOLINE QUINONE, ... | | Authors: | Deng, Y, Ro, S.Y, Rosenzweig, A.C. | | Deposit date: | 2018-05-01 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and function of the lanthanide-dependent methanol dehydrogenase XoxF from the methanotroph Methylomicrobium buryatense 5GB1C.
J. Biol. Inorg. Chem., 23, 2018
|
|
6DBD
 
 | | Crystal Structure of VHH R326 | | Descriptor: | ACETATE ION, SODIUM ION, nanobody VHH R326 | | Authors: | Brooks, C.L, Toride King, M, Huh, I. | | Deposit date: | 2018-05-03 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Structural basis of VHH-mediated neutralization of the food-borne pathogenListeria monocytogenes.
J. Biol. Chem., 293, 2018
|
|
2FQG
 
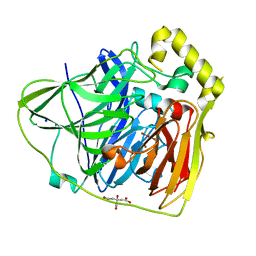 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
6KGH
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis (apo-form) | | Descriptor: | COBALT (II) ION, Penicillin-binding protein PbpB, SODIUM ION | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-11 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KCD
 
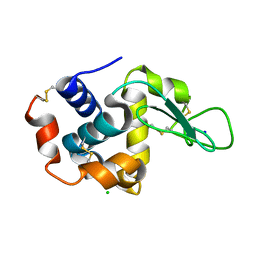 | |
2FWF
 
 | | high resolution crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (reduced form) | | Descriptor: | IODIDE ION, SODIUM ION, Thiol:disulfide interchange protein dsbD | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
6D9D
 
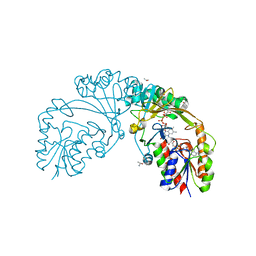 | |
6DFH
 
 | | BG505 MD64 N332-GT2 SOSIP trimer in complex with germline-reverted BG18 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Ozorowski, G, Steichen, J.M, Schief, W.R, Ward, A.B. | | Deposit date: | 2018-05-14 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses.
Science, 366, 2019
|
|
