6XW2
 
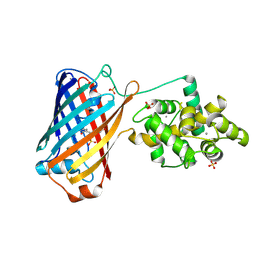 | | Crystal structure of the bright genetically encoded calcium indicator NCaMP7 based on mNeonGreen fluorescent protein | | Descriptor: | CALCIUM ION, Genetically encoded calcium indicator NCaMP7 based on mNeonGreen fluorescent protein, SULFATE ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Lazarenko, V.A, Subach, O.M, Subach, F.V. | | Deposit date: | 2020-01-22 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel Genetically Encoded Bright Positive Calcium Indicator NCaMP7 Based on the mNeonGreen Fluorescent Protein.
Int J Mol Sci, 21, 2020
|
|
2LKW
 
 | |
4UVM
 
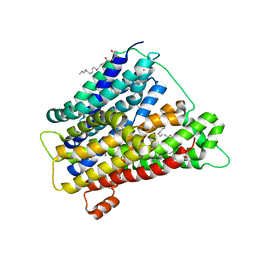 | | In meso crystal structure of the POT family transporter PepTSo | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, GLUTATHIONE UPTAKE TRANSPORTER | | Authors: | Lyons, J.A, Solcan, N, Caffrey, M, Newstead, S. | | Deposit date: | 2014-08-07 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Gating Topology of the Proton-Coupled Oligopeptide Symporters.
Structure, 23, 2015
|
|
7RK9
 
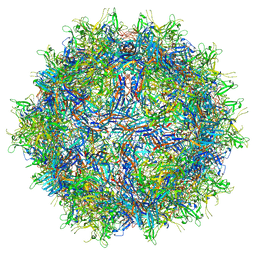 | |
7RK8
 
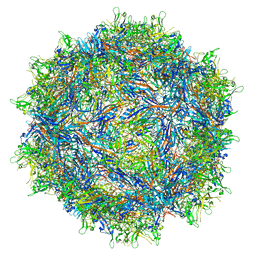 | |
6LU8
 
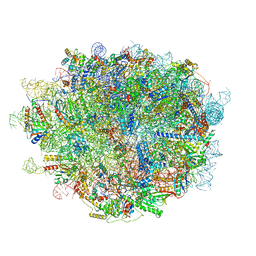 | | Cryo-EM structure of a human pre-60S ribosomal subunit - state A | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Liang, X, Zuo, M, Zhang, Y, Li, N, Ma, C, Dong, M, Gao, N. | | Deposit date: | 2020-01-26 | | Release date: | 2020-08-26 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural snapshots of human pre-60S ribosomal particles before and after nuclear export.
Nat Commun, 11, 2020
|
|
5V6F
 
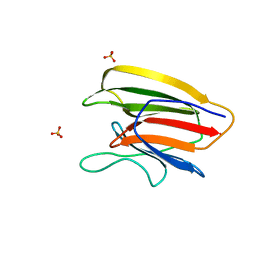 | | Crystal Structure of the Second beta-Prism Domain of RbmC from V. cholerae bound to Mannotriose | | Descriptor: | Hemolysin-related protein, SULFATE ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose | | Authors: | De, S, Kaus, K, Sinclair, S, Case, B.C, Olson, R. | | Deposit date: | 2017-03-16 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of mammalian glycan targeting by Vibrio cholerae cytolysin and biofilm proteins.
PLoS Pathog., 14, 2018
|
|
5V6C
 
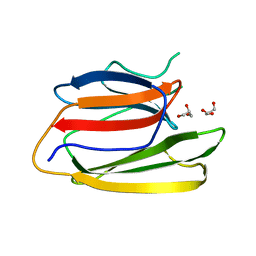 | | Crystal Structure of the Second beta-Prism Domain of RbmC from V. cholerae | | Descriptor: | GLYCEROL, Hemolysin-related protein | | Authors: | De, S, Kaus, K, Sinclair, S, Case, B.C, Olson, R. | | Deposit date: | 2017-03-16 | | Release date: | 2018-01-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of mammalian glycan targeting by Vibrio cholerae cytolysin and biofilm proteins.
PLoS Pathog., 14, 2018
|
|
5VQI
 
 | |
5V6K
 
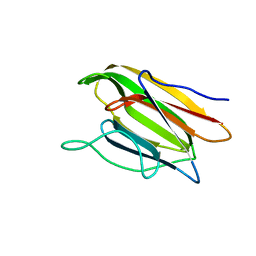 | | Crystal Structure of the Second beta-Prism Domain of RbmC from V. cholerae Bound to N-acetylglucosaminyl-beta-1,2-mannose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, GLYCEROL, Hemolysin-related protein | | Authors: | De, S, Kaus, K, Sinclair, S, Case, B.C, Olson, R. | | Deposit date: | 2017-03-16 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of mammalian glycan targeting by Vibrio cholerae cytolysin and biofilm proteins.
PLoS Pathog., 14, 2018
|
|
1HI7
 
 | |
7TGR
 
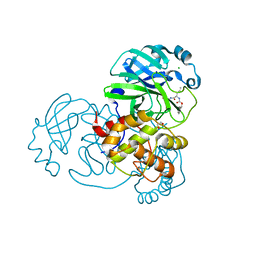 | | Structure of SARS-CoV-2 main protease in complex with GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 1,2-ETHANEDIOL, ... | | Authors: | Esler, M.A, Shi, K, Aihara, H, Harris, R.S. | | Deposit date: | 2022-01-09 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Gain-of-Signal Assays for Probing Inhibition of SARS-CoV-2 M pro /3CL pro in Living Cells.
Mbio, 13, 2022
|
|
4OWK
 
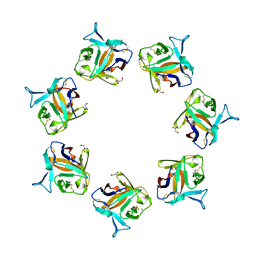 | |
4OWL
 
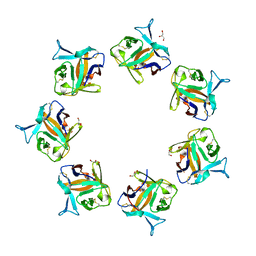 | |
4N0H
 
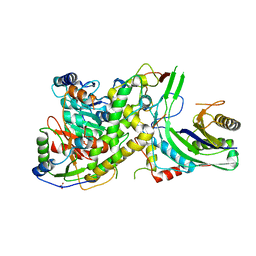 | | Crystal structure of S. cerevisiae mitochondrial GatFAB | | Descriptor: | Glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial, Glutamyl-tRNA(Gln) amidotransferase subunit B, ... | | Authors: | Araiso, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-10-02 | | Release date: | 2014-04-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae mitochondrial GatFAB reveals a novel subunit assembly in tRNA-dependent amidotransferases
Nucleic Acids Res., 42, 2014
|
|
4N0I
 
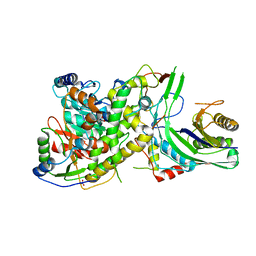 | | Crystal structure of S. cerevisiae mitochondrial GatFAB in complex with glutamine | | Descriptor: | GLUTAMINE, Glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial, ... | | Authors: | Araiso, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-10-02 | | Release date: | 2014-04-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae mitochondrial GatFAB reveals a novel subunit assembly in tRNA-dependent amidotransferases
Nucleic Acids Res., 42, 2014
|
|
4OWJ
 
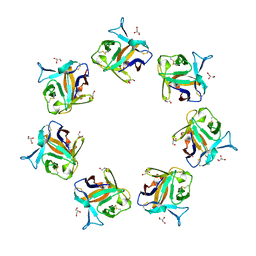 | |
9QH3
 
 | |
9U3R
 
 | | Cryo-EM structure of human AC9 | | Descriptor: | Adenylate cyclase type 9,Protein M2-1 | | Authors: | Suzuki, S, Nomura, R, Suzuki, H, Nishikawa, K, Fujiyoshi, Y. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Activation mechanism of human adenylyl cyclase 9 observed by cryo-electron microscopy
To Be Published
|
|
9U3V
 
 | | Cryo-EM structure of human AC9_delC (TM domain) | | Descriptor: | Adenylate cyclase type 9,Protein M2-1 | | Authors: | Suzuki, S, Nomura, R, Suzuki, H, Nishikawa, K, Fujiyoshi, Y. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Activation mechanism of human adenylyl cyclase 9 observed by cryo-electron microscopy
To Be Published
|
|
9U3P
 
 | | Cryo-EM structure of human AC9-Gs complex (soluble domain) | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Adenylate cyclase type 9,Protein M2-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Suzuki, S, Nomura, R, Suzuki, H, Nishikawa, K, Fujiyoshi, Y. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Activation mechanism of human adenylyl cyclase 9 observed by cryo-electron microscopy
To Be Published
|
|
9U3S
 
 | | Cryo-EM structure of human AC9 (TM domain) | | Descriptor: | Adenylate cyclase type 9,Protein M2-1 | | Authors: | Suzuki, S, Nomura, R, Suzuki, H, Nishikawa, K, Fujiyoshi, Y. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Activation mechanism of human adenylyl cyclase 9 observed by cryo-electron microscopy
To Be Published
|
|
9U3U
 
 | | Cryo-EM structure of human AC9_delC | | Descriptor: | Adenylate cyclase type 9,Protein M2-1 | | Authors: | Suzuki, S, Nomura, R, Suzuki, H, Nishikawa, K, Fujiyoshi, Y. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Activation mechanism of human adenylyl cyclase 9 observed by cryo-electron microscopy
To Be Published
|
|
9U3Q
 
 | | Cryo-EM structure of human AC9-Gs complex (TM domain) | | Descriptor: | Adenylate cyclase type 9,Protein M2-1 | | Authors: | Suzuki, S, Nomura, R, Suzuki, H, Nishikawa, K, Fujiyoshi, Y. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Activation mechanism of human adenylyl cyclase 9 observed by cryo-electron microscopy
To Be Published
|
|
8F1D
 
 | | Voltage-gated potassium channel Kv3.1 apo | | Descriptor: | CHOLESTEROL, POTASSIUM ION, Potassium voltage-gated channel subfamily C member 1, ... | | Authors: | Chen, Y, Ishchenko, A. | | Deposit date: | 2022-11-04 | | Release date: | 2023-10-25 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Identification and structural and biophysical characterization of a positive modulator of human Kv3.1 channels.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
