3JWD
 
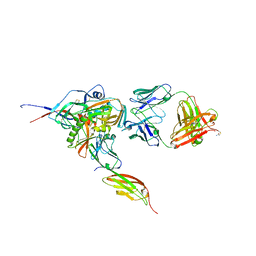 | | Structure of HIV-1 gp120 with gp41-Interactive Region: Layered Architecture and Basis of Conformational Mobility | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB 48D HEAVY CHAIN, FAB 48D LIGHT CHAIN, ... | | Authors: | Pancera, M, Majeed, S, Ban, Y.A, Chen, L, Huang, C.C, Kong, L, Kwon, Y.D, Stuckey, J, Zhou, T, Robinson, J.E, Schief, W.R, Sodroski, J, Wyatt, R, Kwong, P.D. | | Deposit date: | 2009-09-18 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7ADD
 
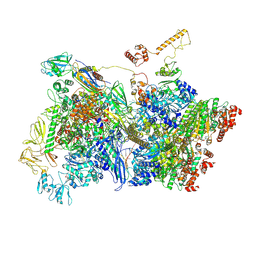 | | Transcription termination intermediate complex IIIa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, C.M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-25 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
7AE3
 
 | | Cryo-EM structure of human RNA Polymerase III elongation complex 3 | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AOV
 
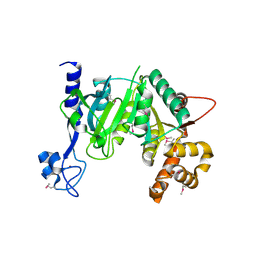 | | Crystal Structure of a TRPM2 Domain | | Descriptor: | Transient receptor potential cation channel subfamily M member 2, ZINC ION | | Authors: | Sander, S, Tidow, H. | | Deposit date: | 2020-10-15 | | Release date: | 2022-03-02 | | Last modified: | 2022-08-17 | | Method: | X-RAY DIFFRACTION (2.00001621 Å) | | Cite: | The crystal structure of TRPM2 MHR1/2 domain reveals a conserved Zn 2+ -binding domain essential for structural integrity and channel activity.
Protein Sci., 31, 2022
|
|
7ADE
 
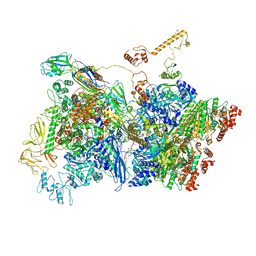 | | Transcription termination complex IVa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, C.M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
7AEA
 
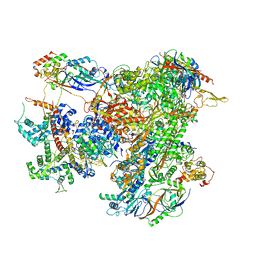 | | Cryo-EM structure of human RNA Polymerase III elongation complex 2 | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3JWN
 
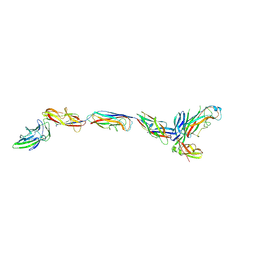 | | Complex of FimC, FimF, FimG and FimH | | Descriptor: | Chaperone protein fimC, FimH protein, GLYCEROL, ... | | Authors: | Le Trong, I, Aprikian, P, Stenkamp, R.E, Sokurenko, E.V. | | Deposit date: | 2009-09-18 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like beta sheet twisting.
Cell(Cambridge,Mass.), 141, 2010
|
|
7A6H
 
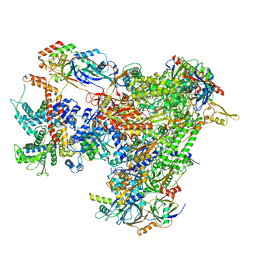 | | Cryo-EM structure of human apo RNA Polymerase III | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-08-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ADB
 
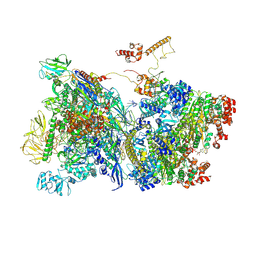 | | Transcription termination intermediate complex 1 delta NusG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, C.M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-04 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
3K1F
 
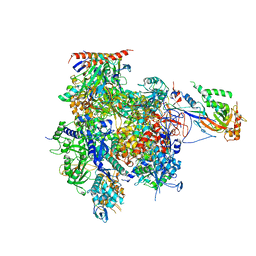 | | Crystal structure of RNA Polymerase II in complex with TFIIB | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Kostrewa, D, Zeller, M.E, Armache, K.-J, Seizl, M, Leike, K, Thomm, M, Cramer, P. | | Deposit date: | 2009-09-27 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | RNA polymerase II-TFIIB structure and mechanism of transcription initiation.
Nature, 462, 2009
|
|
3M66
 
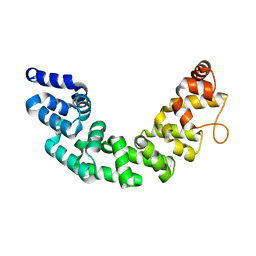 | | Crystal structure of human Mitochondrial Transcription Termination Factor 3 | | Descriptor: | mTERF domain-containing protein 1, mitochondrial | | Authors: | Spahr, H, Samuelsson, T, Hallberg, B.M, Gustafsson, C.M. | | Deposit date: | 2010-03-15 | | Release date: | 2010-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of mitochondrial transcription termination factor 3 reveals a novel nucleic acid-binding domain.
Biochem.Biophys.Res.Commun., 397, 2010
|
|
3MJG
 
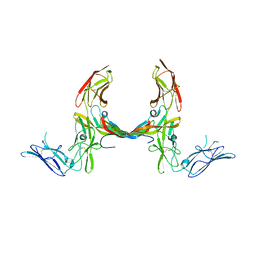 | | The structure of a platelet derived growth factor receptor complex | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-type platelet-derived growth factor receptor, ... | | Authors: | Shim, A.H.R, He, X. | | Deposit date: | 2010-04-12 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of a platelet-derived growth factor/propeptide complex and a platelet-derived growth factor/receptor complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3M4A
 
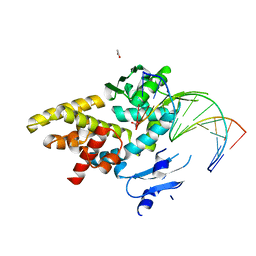 | | Crystal structure of a bacterial topoisomerase IB in complex with DNA reveals a secondary DNA binding site | | Descriptor: | ACETIC ACID, DNA (5'-D(*GP*AP*AP*TP*AP*AP*GP*GP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*CP*CP*CP*TP*TP*AP*TP*TP*C)-3'), ... | | Authors: | Patel, A, Yakovleva, L, Shuman, S, Mondragon, A. | | Deposit date: | 2010-03-10 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a bacterial topoisomerase IB in complex with DNA reveals a secondary DNA binding site.
Structure, 18, 2010
|
|
3MJK
 
 | | Structure of a growth factor precursor | | Descriptor: | Platelet-derived growth factor subunit A | | Authors: | Liu, H, He, X. | | Deposit date: | 2010-04-12 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of a platelet-derived growth factor/propeptide complex and a platelet-derived growth factor/receptor complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3N4S
 
 | | Structure of Csm1 C-terminal domain, P21212 form | | Descriptor: | Monopolin complex subunit CSM1, PENTAETHYLENE GLYCOL | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-22 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
3N4R
 
 | | Structure of Csm1 C-terminal domain, R3 form | | Descriptor: | MALONATE ION, Monopolin complex subunit CSM1, PENTAETHYLENE GLYCOL | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-22 | | Release date: | 2010-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
3N7N
 
 | | Structure of Csm1/Lrs4 complex | | Descriptor: | Monopolin complex subunit CSM1, Monopolin complex subunit LRS4 | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-27 | | Release date: | 2010-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
3NR5
 
 | | Crystal structure of human Maf1 | | Descriptor: | Repressor of RNA polymerase III transcription MAF1 homolog | | Authors: | Ringel, R, Vannini, A, Kusser, A.G, Berninghausen, O, Kassavetis, G.A, Cramer, P. | | Deposit date: | 2010-06-30 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular Basis of RNA Polymerase III Transcription Repression by Maf1
Cell(Cambridge,Mass.), 143, 2010
|
|
4NZF
 
 | | Crystal structure of Abp-D197A (a GH27-b-L-arabinopyranosidase from Geobacillus stearothermophilus), in complex with arabinose | | Descriptor: | Abp, a GH27 beta-L-arabinopyranosidase, CITRIC ACID, ... | | Authors: | Lansky, S, Solomon, H.V, Salama, R, Belrhali, H, Shoham, Y, Shoham, G. | | Deposit date: | 2013-12-12 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-specificity relationships in Abp, a GH27 beta-L-arabinopyranosidase from Geobacillus stearothermophilus T6
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3OLC
 
 | |
3P91
 
 | |
3P22
 
 | | Crystal structure of the ENE, a viral RNA stability element, in complex with A9 RNA | | Descriptor: | Core ENE hairpin from KSHV PAN RNA, oligo(A)9 RNA | | Authors: | Mitton-Fry, R.M, DeGregorio, S.J, Wang, J, Steitz, T.A, Steitz, J.A. | | Deposit date: | 2010-10-01 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Poly(A) tail recognition by a viral RNA element through assembly of a triple helix.
Science, 330, 2010
|
|
3ORY
 
 | | Crystal structure of Flap endonuclease 1 from hyperthermophilic archaeon Desulfurococcus amylolyticus | | Descriptor: | PHOSPHATE ION, flap endonuclease 1 | | Authors: | Mase, T, Kubota, K, Miyazono, K, Kawarabayashii, Y, Tanokura, M. | | Deposit date: | 2010-09-08 | | Release date: | 2011-02-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of flap endonuclease 1 from the hyperthermophilic archaeon Desulfurococcus amylolyticus
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3P8B
 
 | |
4OWR
 
 | | Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair Rae1-Nup98 | | Descriptor: | Matrix protein, Nuclear pore complex protein Nup98-Nup96, mRNA export factor | | Authors: | Ren, Y, Quan, B, Seo, H.S, Blobel, G. | | Deposit date: | 2014-02-03 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair (Rae1 Nup98).
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
