3KES
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae in the Hexagonal, P61 space group | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
7Q1E
 
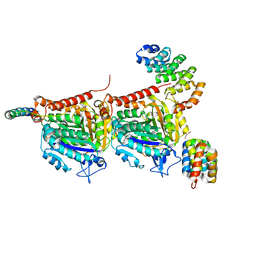 | | CPAP:TUBULIN:IIH5 ALPHAREP COMPLEX | | Descriptor: | Centromere protein J, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Campanacci, V, Gigant, b. | | Deposit date: | 2021-10-19 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural convergence for tubulin binding of CPAP and vinca domain microtubule inhibitors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DJ6
 
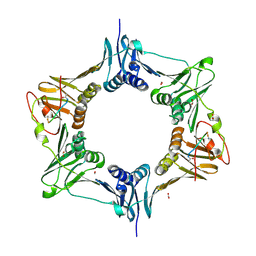 | | Sliding-clamp-ImuB peptide | | Descriptor: | ACETATE ION, Beta sliding clamp, FORMIC ACID, ... | | Authors: | Kapur, M.K, Gray, O.J, Honzatko, R.H, Nelson, S.N. | | Deposit date: | 2022-06-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interaction of sliding clamp with mycobacterial polymerases
To Be Published
|
|
7Q1F
 
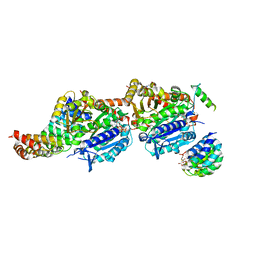 | | CPAP:TUBULIN:IE5 ALPHAREP COMPLEX | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CITRATE ANION, Centromere protein J, ... | | Authors: | Campanacci, V, Gigant, b. | | Deposit date: | 2021-10-19 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.347 Å) | | Cite: | Structural convergence for tubulin binding of CPAP and vinca domain microtubule inhibitors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3Q3W
 
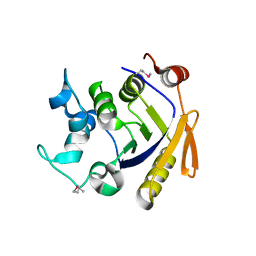 | | Isopropylmalate isomerase small subunit from Campylobacter jejuni. | | Descriptor: | 3-isopropylmalate dehydratase small subunit | | Authors: | Osipiuk, J, Gu, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-22 | | Release date: | 2011-01-26 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Isopropylmalate isomerase small subunit from Campylobacter jejuni.
To be Published
|
|
1DYR
 
 | | THE STRUCTURE OF PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE TO 1.9 ANGSTROMS RESOLUTION | | Descriptor: | DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIMETHOPRIM | | Authors: | Champness, J.N, Achari, A, Ballantine, S.P, Bryant, P.K, Delves, C.J, Stammers, D.K. | | Deposit date: | 1994-09-14 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structure of Pneumocystis carinii dihydrofolate reductase to 1.9 A resolution.
Structure, 2, 1994
|
|
7Q33
 
 | |
3KEP
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
7ZAP
 
 | |
5WDZ
 
 | |
1XNB
 
 | |
1XND
 
 | |
3NF5
 
 | | Crystal structure of the C-terminal domain of nuclear pore complex component NUP116 from Candida glabrata | | Descriptor: | GLYCEROL, Nucleoporin NUP116 | | Authors: | Sampathkumar, P, Manglicmot, D, Bain, K, Gilmore, J, Gheyi, T, Rout, M, Sali, A, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata.
Proteins, 80, 2012
|
|
6BDF
 
 | | 2.8 A resolution reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Campbell, M.G, Veesler, D, Cheng, A, Potter, C.S, Carragher, B. | | Deposit date: | 2017-10-23 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | 2.8 angstrom resolution reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy.
Elife, 4, 2015
|
|
3RFW
 
 | | The virulence factor PEB4 and the periplasmic protein Cj1289 are two structurally-related SurA-like chaperones in the human pathogen Campylobacter jejuni | | Descriptor: | Cell-binding factor 2 | | Authors: | Kale, A, Phansopa, C, Suwannachart, C, Craven, C.J, Rafferty, J, Kelly, D.J. | | Deposit date: | 2011-04-07 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The virulence factor PEB4 and the periplasmic protein Cj1289 are two structurally-related SurA-like chaperones in the human pathogen Campylobacter jejuni
To be Published
|
|
3N7C
 
 | | Crystal structure of the RAN binding domain from the nuclear pore complex component NUP2 from Ashbya gossypii | | Descriptor: | ABR034Wp | | Authors: | Sampathkumar, P, Manglicmot, D, Gilmore, J, Bain, K, Gheyi, T, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-26 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of the RAN binding domain from the nuclear pore complex component NUP2 from Ashbya gossypii
To be Published
|
|
3RGC
 
 | | The virulence factor PEB4 and the periplasmic protein Cj1289 are two structurally related SurA-like chaperones in the human pathogen Campylobacter jejuni | | Descriptor: | Possible periplasmic protein | | Authors: | Kale, A, Phansopa, C, Suwannachart, C, Craven, C.J, Rafferty, J, Kelly, D.J. | | Deposit date: | 2011-04-08 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The virulence factor PEB4 and the periplasmic protein Cj1289 are two structurally related SurA-like chaperones in the human pathogen Campylobacter jejuni
To be Published
|
|
1ESJ
 
 | | CRYSTAL STRUCTURE OF THIAZOLE KINASE MUTANT (C198S) | | Descriptor: | HYDROXYETHYLTHIAZOLE KINASE, SULFATE ION | | Authors: | Campobasso, N, Mathews, I.I, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-04-10 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of 4-methyl-5-beta-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 A resolution.
Biochemistry, 39, 2000
|
|
3NO8
 
 | | Crystal structure of the PHR domain from human BTBD2 Protein | | Descriptor: | BTB/POZ domain-containing protein 2, GLYCEROL, SULFATE ION | | Authors: | Sampathkumar, P, Miller, S, Rutter, M, Bain, K, Gheyi, T, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-24 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the PHR domain from human BTBD2 Protein
To be Published
|
|
3NWA
 
 | | Glycoprotein B from Herpes simplex virus type 1, W174R mutant, low-pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein B, MESO-ERYTHRITOL | | Authors: | Stampfer, S.D, Lou, H, Cohen, G.H, Eisenberg, R.J, Heldwein, E.E. | | Deposit date: | 2010-07-09 | | Release date: | 2010-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2629 Å) | | Cite: | Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1.
J.Virol., 84, 2010
|
|
1ESQ
 
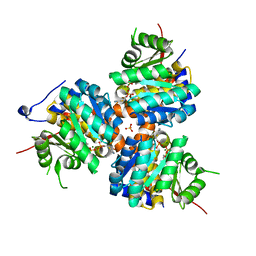 | | CRYSTAL STRUCTURE OF THIAZOLE KINASE MUTANT (C198S) WITH ATP AND THIAZOLE PHOSPHATE. | | Descriptor: | 4-METHYL-5-HYDROXYETHYLTHIAZOLE PHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, HYDROXYETHYLTHIAZOLE KINASE, ... | | Authors: | Campobasso, N, Mathews, I.I, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-04-10 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 4-methyl-5-beta-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 A resolution.
Biochemistry, 39, 2000
|
|
1G1U
 
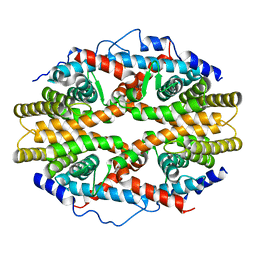 | | THE 2.5 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE RXRALPHA LIGAND BINDING DOMAIN IN TETRAMER IN THE ABSENCE OF LIGAND | | Descriptor: | RETINOIC ACID RECEPTOR RXR-ALPHA | | Authors: | Gampe Jr, R.T, Montana, V.G, Lambert, M.H, Wisely, G.B, Milburn, M.V, Xu, H.E. | | Deposit date: | 2000-10-13 | | Release date: | 2001-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for autorepression of retinoid X receptor by tetramer formation and the AF-2 helix.
Genes Dev., 14, 2000
|
|
1G5Y
 
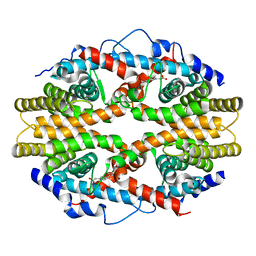 | | THE 2.0 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE RXRALPHA LIGAND BINDING DOMAIN TETRAMER IN THE PRESENCE OF A NON-ACTIVATING RETINOIC ACID ISOMER. | | Descriptor: | RETINOIC ACID, RETINOIC ACID RECEPTOR RXR-ALPHA | | Authors: | Gampe Jr, R.T, Montana, V.G, Lambert, M.H, Wisely, G.B, Milburn, M.V, Xu, H.E. | | Deposit date: | 2000-11-02 | | Release date: | 2001-05-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for autorepression of retinoid X receptor by tetramer formation and the AF-2 helix.
Genes Dev., 14, 2000
|
|
1I0A
 
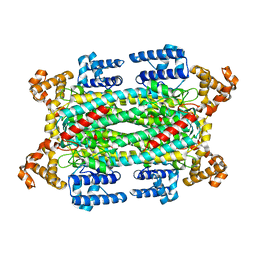 | |
2VD2
 
 | |
