7R4G
 
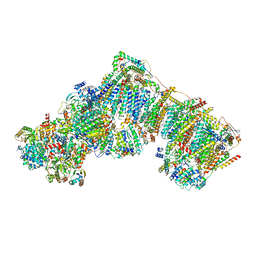 | | Bovine complex I in the presence of IM1761092, slack class ii (Composite map) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1-carbamimidoyl-3-[2-(3-chloranyl-4-iodanyl-phenyl)ethyl]guanidine, ... | | Authors: | Bridges, H.R, Blaza, J.N, Yin, Z, Chung, I, Hirst, J. | | Deposit date: | 2022-02-08 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis of mammalian respiratory complex I inhibition by medicinal biguanides.
Science, 379, 2023
|
|
3DMR
 
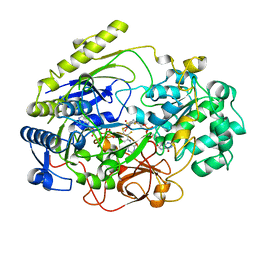 | | STRUCTURE OF DMSO REDUCTASE FROM RHODOBACTER CAPSULATUS AT PH 7.0 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DMSO REDUCTASE, MOLYBDENUM(VI) ION, ... | | Authors: | Mcalpine, A.S, Bailey, S. | | Deposit date: | 1997-04-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molybdenum Active Centre of Dmso Reductase from Rhodobacter Capsulatus: Crystal Structure of the Oxidised Enzyme at 1.82-A Resolution and the Dithionite-Reduced Enzyme at 2.8-A Resolution
J.Biol.Inorg.Chem., 2, 1997
|
|
7P64
 
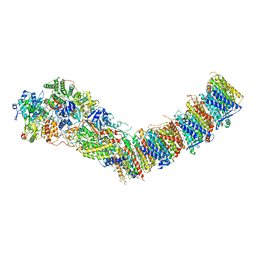 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 6, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
5T5M
 
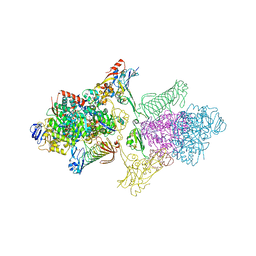 | | TUNGSTEN-CONTAINING FORMYLMETHANOFURAN DEHYDROGENASE FROM METHANOTHERMOBACTER WOLFEII, TRIGONAL FORM AT 2.5 A. | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, GLYCEROL, HYDROSULFURIC ACID, ... | | Authors: | Wagner, T, Ermler, U, Shima, S. | | Deposit date: | 2016-08-31 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The methanogenic CO2 reducing-and-fixing enzyme is bifunctional and contains 46 [4Fe-4S] clusters.
Science, 354, 2016
|
|
7VZ1
 
 | | Matrix arm of deactive state CI from Q1-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-15 | | Release date: | 2022-12-28 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1Y5L
 
 | | The crystal structure of the NarGHI mutant NarI-H66Y | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
1Y5N
 
 | | The crystal structure of the NarGHI mutant NarI-K86A in complex with pentachlorophenol | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
1TMO
 
 | | TRIMETHYLAMINE N-OXIDE REDUCTASE FROM SHEWANELLA MASSILIA | | Descriptor: | GUANYLATE-O'-PHOSPHORIC ACID MONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,5,6,7,8A,9,10,10A-OCTAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL) ESTER, MOLYBDENUM (IV)OXIDE, TRIMETHYLAMINE N-OXIDE REDUCTASE | | Authors: | Czjzek, M, Dos Santos, J.P, Giordano, G, Mejean, V. | | Deposit date: | 1998-08-03 | | Release date: | 1999-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of oxidized trimethylamine N-oxide reductase from Shewanella massilia at 2.5 A resolution.
J.Mol.Biol., 284, 1998
|
|
7TGH
 
 | | Cryo-EM structure of respiratory super-complex CI+III2 from Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2 iron, ... | | Authors: | Zhou, L, Maldonado, M, Padavannil, A, Guo, F, Letts, J.A. | | Deposit date: | 2022-01-07 | | Release date: | 2022-04-06 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of Tetrahymena 's respiratory chain reveal the diversity of eukaryotic core metabolism.
Science, 376, 2022
|
|
7V2R
 
 | | Active state complex I from Q1-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V33
 
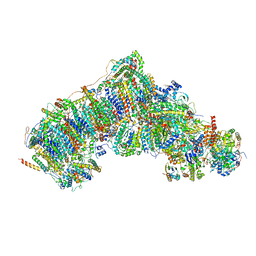 | | Active state complex I from rotenone-NADH dataset | | Descriptor: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8E9G
 
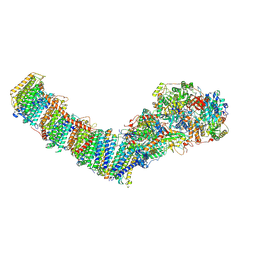 | | Mycobacterial respiratory complex I with both quinone positions modelled | | Descriptor: | (2R)-3-{[(R)-hydroxy({(1S,2R,3R,4R,5S,6S)-3,4,5-trihydroxy-2-(alpha-D-mannopyranosyloxy)-6-[(6-O-undecanoyl-beta-D-mannopyranosyl)oxy]cyclohexyl}oxy)phosphoryl]oxy}-2-(octanoyloxy)propyl undecanoate, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liang, Y, Rubinstein, J.L. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-12 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of mycobacterial respiratory complex I.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7VWJ
 
 | | Matrix arm of deactive state CI from rotenone-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-10 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VY8
 
 | | Matrix arm of active state CI from Q10-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-13 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W1Z
 
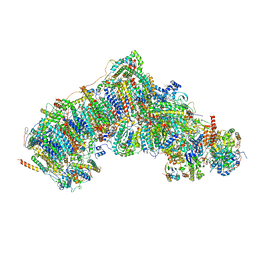 | | Active state CI from Rotenone-NADH dataset, Subclass 2 | | Descriptor: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-21 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W2U
 
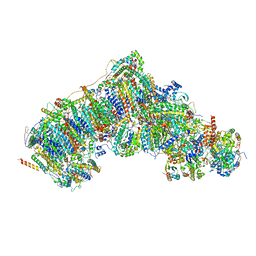 | | Active state CI from DQ-NADH dataset, Subclass 2 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-24 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZCI
 
 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-28 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
6YJ4
 
 | | Structure of Yarrowia lipolytica complex I at 2.7 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Acyl carrier protein ACPM1 of NADH:Ubiquinone Oxidoreductase (Complex I), Acyl carrier protein ACPM2 of NADH:Ubiquinone Oxidoreductase (Complex I), ... | | Authors: | Hirst, J, Grba, D. | | Deposit date: | 2020-04-02 | | Release date: | 2020-08-12 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mitochondrial complex I structure reveals ordered water molecules for catalysis and proton translocation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7V2K
 
 | | Deactive state complex I from DQ-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V30
 
 | | Deactive state complex I from Q1-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8E9H
 
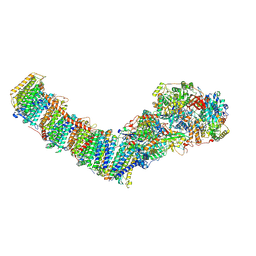 | | Mycobacterial respiratory complex I, fully-inserted quinone | | Descriptor: | (2R)-3-{[(R)-hydroxy({(1S,2R,3R,4R,5S,6S)-3,4,5-trihydroxy-2-(alpha-D-mannopyranosyloxy)-6-[(6-O-undecanoyl-beta-D-mannopyranosyl)oxy]cyclohexyl}oxy)phosphoryl]oxy}-2-(octanoyloxy)propyl undecanoate, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liang, Y, Rubinstein, J.L. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-12 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of mycobacterial respiratory complex I.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7P7E
 
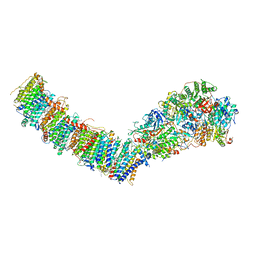 | | Complex I from E. coli, DDM/LMNG-purified, Apo, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P7J
 
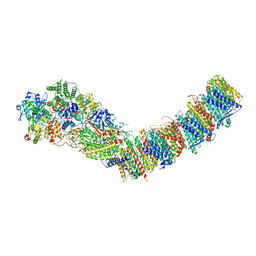 | | Complex I from E. coli, DDM/LMNG-purified, with DQ, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7VXP
 
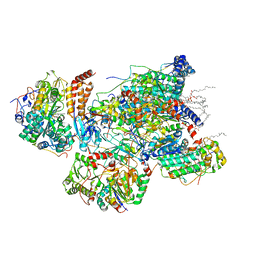 | | Matrix arm of active state CI from Q10 dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Acyl carrier protein, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-13 | | Release date: | 2022-11-23 | | Last modified: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W4F
 
 | | Active state CI from Q1-NADH dataset, Subclass 4 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-27 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
