6ZJN
 
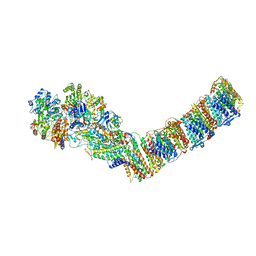 | | Respiratory complex I from Thermus thermophilus, NADH dataset, minor state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, NADH-quinone oxidoreductase subunit 1, ... | | Authors: | Kaszuba, K, Tambalo, M, Gallagher, G.T, Sazanov, L.A. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
6ZJY
 
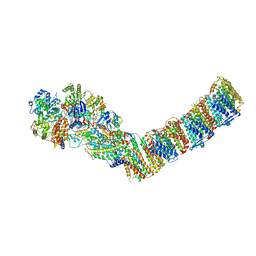 | | Respiratory complex I from Thermus thermophilus, NAD+ dataset, minor state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, NADH-quinone oxidoreductase subunit 1, ... | | Authors: | Kaszuba, K, Tambalo, M, Gallagher, G.T, Sazanov, L.A. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
6ZR2
 
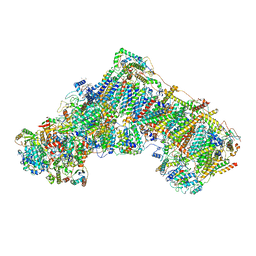 | | Cryo-EM structure of respiratory complex I in the active state from Mus musculus at 3.1 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bridges, H.R, Blaza, J.N, Agip, A.N.A, Hirst, J. | | Deposit date: | 2020-07-10 | | Release date: | 2020-10-21 | | Last modified: | 2020-10-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of inhibitor-bound mammalian complex I.
Nat Commun, 11, 2020
|
|
6ZTQ
 
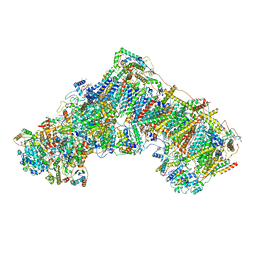 | | Cryo-EM structure of respiratory complex I from Mus musculus inhibited by piericidin A at 3.0 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bridges, H.R, Blaza, J.N, Agip, A.N.A, Hirst, J. | | Deposit date: | 2020-07-20 | | Release date: | 2020-10-21 | | Last modified: | 2020-10-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of inhibitor-bound mammalian complex I.
Nat Commun, 11, 2020
|
|
7A23
 
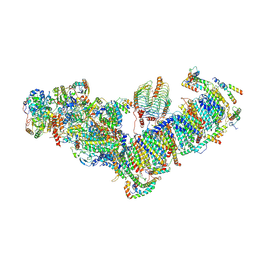 | | Plant mitochondrial respiratory complex I | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 13kDa, 15kDa, ... | | Authors: | Soufari, H, Waltz, F, Hashem, Y. | | Deposit date: | 2020-08-15 | | Release date: | 2020-10-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Specific features and assembly of the plant mitochondrial complex I revealed by cryo-EM.
Nat Commun, 11, 2020
|
|
7A24
 
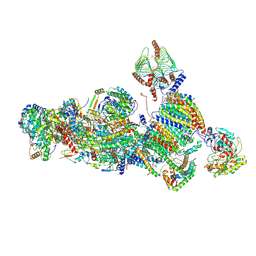 | | Assembly intermediate of the plant mitochondrial complex I | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 13kDa, 15kDa, ... | | Authors: | Soufari, H, Waltz, F, Hashem, Y. | | Deposit date: | 2020-08-16 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Specific features and assembly of the plant mitochondrial complex I revealed by cryo-EM.
Nat Commun, 11, 2020
|
|
7AK5
 
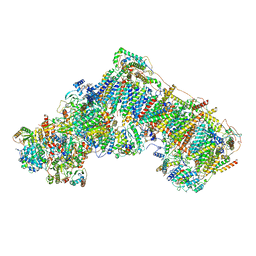 | | Cryo-EM structure of respiratory complex I in the deactive state from Mus musculus at 3.2 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yin, Z, Bridges, H.R, Grba, D, Hirst, J. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural basis for a complex I mutation that blocks pathological ROS production.
Nat Commun, 12, 2021
|
|
7AK6
 
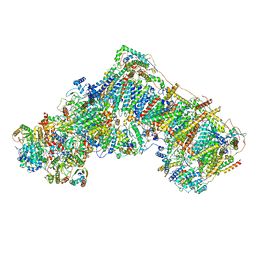 | | Cryo-EM structure of ND6-P25L mutant respiratory complex I from Mus musculus at 3.8 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yin, Z, Bridges, H.R, Grba, D, Hirst, J. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for a complex I mutation that blocks pathological ROS production.
Nat Commun, 12, 2021
|
|
7AQR
 
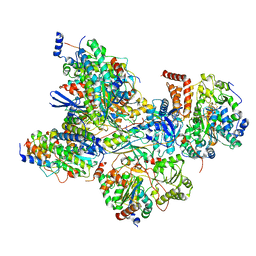 | | Cryo-EM structure of Arabidopsis thaliana Complex-I (peripheral arm) | | Descriptor: | Acyl carrier protein 2, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-22 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7AR7
 
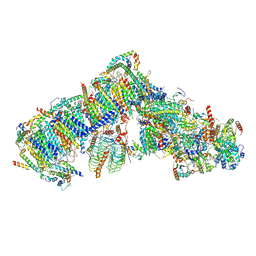 | | Cryo-EM structure of Arabidopsis thaliana complex-I (open conformation) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, ... | | Authors: | Klusch, N, Kuelbrandt, W. | | Deposit date: | 2020-10-23 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7AR8
 
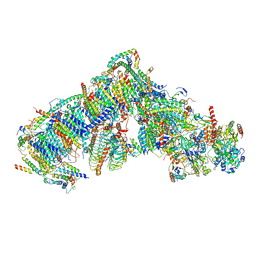 | | Cryo-EM structure of Arabidopsis thaliana complex-I (closed conformation) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, ... | | Authors: | Klusch, N, Kuelbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7ARB
 
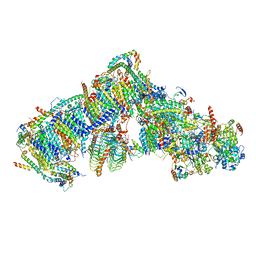 | | Cryo-EM structure of Arabidopsis thaliana Complex-I (complete composition) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, ... | | Authors: | Klusch, N, Kuelbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7AWT
 
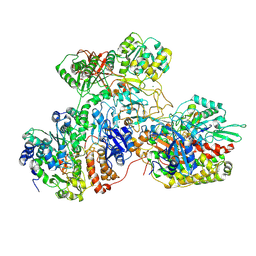 | | E. coli NADH quinone oxidoreductase hydrophilic arm | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Schimpf, J, Grishkovskaya, I, Haselbach, D, Friedrich, T. | | Deposit date: | 2020-11-09 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structure of the peripheral arm of a minimalistic respiratory complex I.
Structure, 30, 2022
|
|
7B04
 
 | | Structure of Nitrite oxidoreductase (Nxr) from the anammox bacterium Kuenenia stuttgartiensis. | | Descriptor: | CALCIUM ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Moreno-Chicano, T, Dietl, A, Akram, M, Barends, T.R.M. | | Deposit date: | 2020-11-18 | | Release date: | 2021-07-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural and functional characterization of the intracellular filament-forming nitrite oxidoreductase multiprotein complex
Nat Microbiol, 2021
|
|
7B0N
 
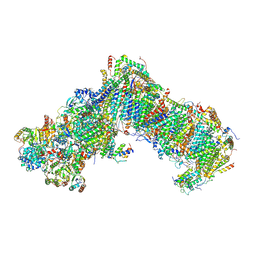 | |
7B93
 
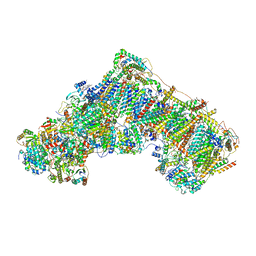 | | Cryo-EM structure of mitochondrial complex I from Mus musculus inhibited by IACS-2858 at 3.0 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1-[[3-(4-methylsulfonylpiperidin-1-yl)phenyl]methyl]-5-[3-[4-(trifluoromethyloxy)phenyl]-1,2,4-oxadiazol-5-yl]pyridin-2-one, ... | | Authors: | Chung, I, Hirst, J. | | Deposit date: | 2020-12-14 | | Release date: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Cork-in-bottle mechanism of inhibitor binding to mammalian complex I.
Sci Adv, 7, 2021
|
|
7BKB
 
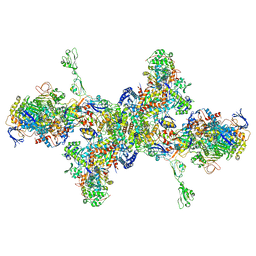 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (hexameric, composite structure) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
7BKC
 
 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (dimeric, composite structure) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
7BKD
 
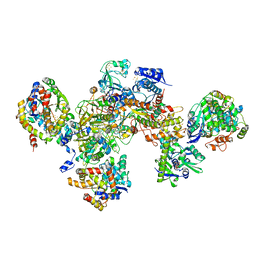 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (heterodislfide reductase core and mobile arm in conformational state 1, composite structure) | | Descriptor: | CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, CoB--CoM heterodisulfide reductase subunit C, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
7BKE
 
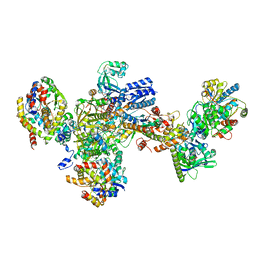 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (heterodisulfide reductase core and mobile arm in conformational state 2, composite structure) | | Descriptor: | CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, CoB--CoM heterodisulfide reductase subunit C, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
7DGQ
 
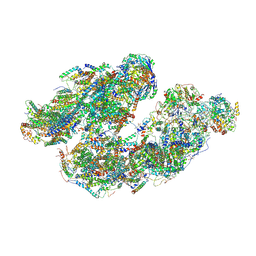 | | Activity optimized supercomplex state1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DGR
 
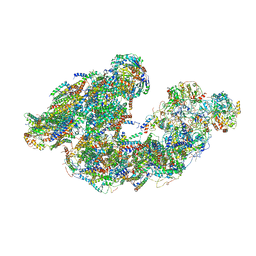 | | Activity optimized supercomplex state2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DGS
 
 | | Activity optimized supercomplex state3 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DGZ
 
 | | Activity optimized complex I (closed form) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DH0
 
 | | Activity optimized complex I (open form) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
