2KFP
 
 | | Solution NMR structure of PSPTO_3016 from Pseudomonas syringae. Northeast Structural Genomics Consortium target PsR293. | | Descriptor: | PSPTO_3016 protein | | Authors: | Feldmann, E.A, Ramelot, T.A, Zhao, L, Hamilton, K, Ciccosanti, C, Xiao, R, Nair, R, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR and X-ray crystal structures of Pseudomonas syringae Pspto_3016 from protein domain family PF04237 (DUF419) adopt a "double wing" DNA binding motif.
J.Struct.Funct.Genom., 13, 2012
|
|
2A6M
 
 | | Crystal Structure of the ISHp608 Transposase | | Descriptor: | ISHp608 transposase | | Authors: | Ronning, D.R, Guynet, C, Ton-Hoang, B, Perez, Z.N, Ghirlando, R, Chandler, M, Dyda, F. | | Deposit date: | 2005-07-03 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Active site sharing and subterminal hairpin recognition in a new class of DNA transposases.
Mol.Cell, 20, 2005
|
|
1MC8
 
 | | Crystal Structure of Flap Endonuclease-1 R42E mutant from Pyrococcus horikoshii | | Descriptor: | Flap Endonuclease-1 | | Authors: | Matsui, E, Musti, K.V, Abe, J, Yamazaki, K, Matsui, I, Harata, K. | | Deposit date: | 2002-08-06 | | Release date: | 2002-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Structure and Novel DNA Binding Sites Located in Loops of Flap Endonuclease-1 from Pyrococcus horikoshii
J.BIOL.CHEM., 277, 2002
|
|
6IET
 
 | | The crystal structure of TRIM66 PHD-Bromo domain | | Descriptor: | Tripartite motif-containing protein 66, ZINC ION | | Authors: | Chen, J. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | TRIM66 reads unmodified H3R2K4 and H3K56ac to respond to DNA damage in embryonic stem cells.
Nat Commun, 10, 2019
|
|
3GJK
 
 | | crystal structure of a DNA duplex containing 7,8-dihydropyridol[2,3-d]pyrimidin-2-one | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*A)-3', 5'-D(P*TP*TP*(B7C)P*GP*CP*G)-3', POTASSIUM ION | | Authors: | Takenaka, A, Juan, E.C.M, Shimizu, S, Haraguchi, T, Xiao, M, Kurose, T, Ohkubo, A, Sekine, M, Shibata, T, Millington, C.L, Williams, D.M. | | Deposit date: | 2009-03-09 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into the stabilizing contributions of bicyclic cytosine analogues: crystal structures of DNA duplexes containing 7,8-dihydropyridol[2,3-d]pyrimidin-2-one
To be Published
|
|
2GBZ
 
 | | The Crystal Structure of XC847 from Xanthomonas campestris: a 3-5 Oligoribonuclease of DnaQ fold family with a Novel Opposingly-Shifted Helix | | Descriptor: | MAGNESIUM ION, Oligoribonuclease | | Authors: | Chin, K.H, Yang, C.Y, Chou, C.C, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2006-03-12 | | Release date: | 2007-01-16 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of XC847 from Xanthomonas campestris: a 3'-5' oligoribonuclease of DnaQ fold family with a novel opposingly shifted helix
Proteins, 65, 2006
|
|
1WEG
 
 | | Catalytic Domain Of Muty From Escherichia Coli K142A Mutant | | Descriptor: | 1,2-ETHANEDIOL, A/G-specific adenine glycosylase, IMIDAZOLE, ... | | Authors: | Hitomi, K, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2004-05-25 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reaction intermediates in the catalytic mechanism of Escherichia coli MutY DNA glycosylase
J.Biol.Chem., 279, 2004
|
|
3GJL
 
 | | crystal structure of a DNA duplex containing 7,8-dihydropyridol[2,3-d]pyrimidin-2-one | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*A)-3', 5'-D(P*TP*TP*(B7C)P*GP*CP*G)-3', SODIUM ION | | Authors: | Takenaka, A, Juan, E.C.M, Shimizu, S, Haraguchi, T, Xiao, M, Kurose, T, Ohkubo, A, Sekine, M, Shibata, T, Millington, C.L, Williams, D.M. | | Deposit date: | 2009-03-09 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Insights into the stabilizing contributions of bicyclic cytosine analogues: crystal structures of DNA duplexes containing 7,8-dihydropyridol[2,3-d]pyrimidin-2-one
To be Published
|
|
1WEF
 
 | |
1WEI
 
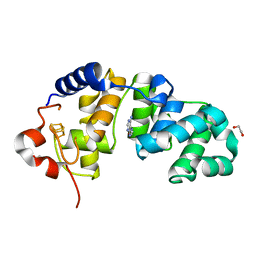 | | Catalytic Domain Of Muty From Escherichia Coli K20A Mutant Complexed To Adenine | | Descriptor: | 1,2-ETHANEDIOL, A/G-specific adenine glycosylase, ADENINE, ... | | Authors: | Hitomi, K, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2004-05-25 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Reaction intermediates in the catalytic mechanism of Escherichia coli MutY DNA glycosylase
J.Biol.Chem., 279, 2004
|
|
6H09
 
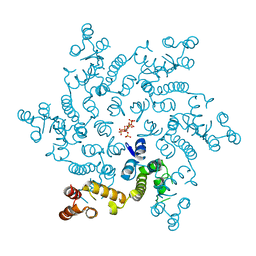 | | HIV capsid hexamer with IP6 ligand | | Descriptor: | Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | James, L.C. | | Deposit date: | 2018-07-06 | | Release date: | 2018-08-15 | | Last modified: | 2020-10-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis.
Elife, 7, 2018
|
|
1NGT
 
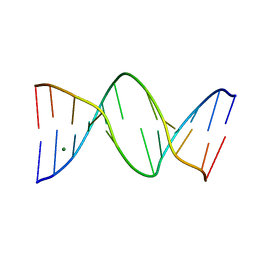 | | The Role of Minor Groove Functional Groups in DNA Hydration | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*(MTR)P*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Woods, K.K, Lan, T, McLaughlin, L.W, Williams, L.D. | | Deposit date: | 2002-12-17 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Role of Minor Groove Functional Groups in DNA Hydration
Nucleic Acids Res., 31, 2003
|
|
7CUX
 
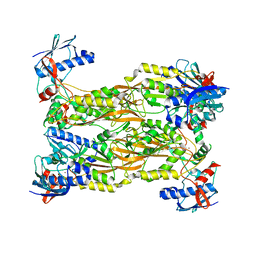 | | Crystal structure of human Schlafen 5 N'-terminal domain (SLFN5-N) involved in ssRNA cleaving and DNA binding | | Descriptor: | Schlafen family member 5, ZINC ION | | Authors: | Yang, J.Y, Luo, M, Ou, J.Y, Wang, Z.W, Gao, S. | | Deposit date: | 2020-08-25 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.29477072 Å) | | Cite: | Crystal structure of human Schlafen 5 N'-terminal domain (SLFN5-N) involved in ssRNA cleaving and DNA binding
To Be Published
|
|
1K9L
 
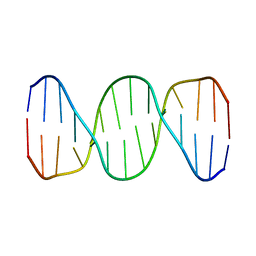 | |
1K5F
 
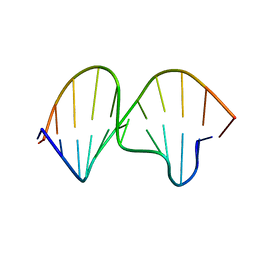 | | SOLUTION STRUCTURE OF THE S-STYRENE ADDUCT IN THE RAS61 SEQUENCE | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(ABS)P*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3' | | Authors: | Hennard, C, Finneman, J, Harris, C.M, Harris, T.M, Stone, M.P. | | Deposit date: | 2001-10-10 | | Release date: | 2002-01-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The nonmutagenic (R)- and (S)-beta-(N(6)-adenyl)styrene oxide adducts are oriented in the major groove and show little perturbation to DNA
structure.
Biochemistry, 40, 2001
|
|
6IEU
 
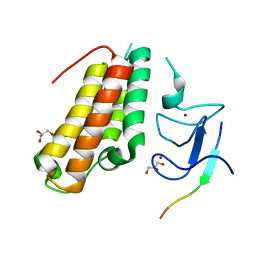 | |
1PVE
 
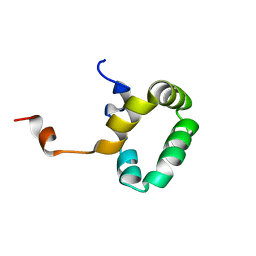 | |
1K5E
 
 | | Solution Structure of R-styrene Adduct in the Ras61 Sequence | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(ABR)P*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3' | | Authors: | Hennard, C, Finneman, J, Harris, C.M, Harris, T.M, Stone, M.P. | | Deposit date: | 2001-10-10 | | Release date: | 2002-01-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The nonmutagenic (R)- and (S)-beta-(N(6)-adenyl)styrene oxide adducts are oriented in the major groove and show little perturbation to DNA
structure.
Biochemistry, 40, 2001
|
|
1K9H
 
 | | NMR structure of DNA TGTGAGCGCTCACA | | Descriptor: | 5'-D(*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*A)-3' | | Authors: | Kaluarachchi, K, Gorenstein, D.G, Luxon, B.A. | | Deposit date: | 2001-10-29 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | How Do Proteins Recognize DNA? Solution Structure and Local Conformational Dynamics of Lac Operators by 2D NMR
J.Biomol.Struct.Dyn., Conversation 11, 2000
|
|
2OIH
 
 | | Hepatitis Delta Virus gemonic ribozyme precursor with C75U mutation and bound to monovalent cation Tl+ | | Descriptor: | HDV ribozyme, THALLIUM (I) ION, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Ding, F, Batchelor, J.D, Doudna, J.A. | | Deposit date: | 2007-01-11 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural roles of monovalent cations in the HDV ribozyme.
Structure, 15, 2007
|
|
2OJ3
 
 | | Hepatitis Delta Virus ribozyme precursor structure, with C75U mutation, bound to Tl+ and cobalt hexammine (Co(NH3)63+) | | Descriptor: | COBALT HEXAMMINE(III), HDV RIBOZYME, THALLIUM (I) ION, ... | | Authors: | Ke, A, Ding, F, Batchelor, J.D, Doudna, J.A. | | Deposit date: | 2007-01-12 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural roles of monovalent cations in the HDV ribozyme.
Structure, 15, 2007
|
|
1L4J
 
 | | Holliday Junction TCGGTACCGA with Na and Ca Binding Sites. | | Descriptor: | 5'-D(*TP*CP*GP*GP*TP*AP*CP*CP*GP*A)-3', CALCIUM ION, SODIUM ION | | Authors: | Thorpe, J.H, Gale, B.C, Teixeira, S.C.M, Cardin, C.J. | | Deposit date: | 2002-03-05 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational and Hydration Effects of Site-selective Sodium, Calcium and
Strontium Ion Binding to the DNA Holliday Junction Structure
d(TCGGTACCGA)4
J.Mol.Biol., 327, 2003
|
|
3H5K
 
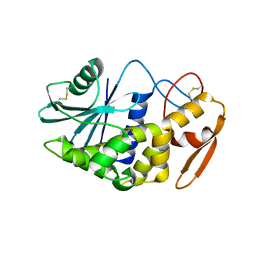 | | Crystal structure of the ribosome inactivating protein PDL1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome-inactivating protein PD-L1/PD-L2 | | Authors: | Ruggiero, A, Di Maro, A, Severino, V, Chambery, A, Berisio, R. | | Deposit date: | 2009-04-22 | | Release date: | 2009-10-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of PD-L1, a ribosome inactivating protein from Phytolacca dioica L. Leaves with the property to induce DNA cleavage
Biopolymers, 91, 2009
|
|
3KDK
 
 | |
3KDG
 
 | |
