5C05
 
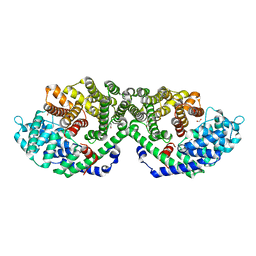 | | Crystal Structure of Gamma-terpinene Synthase from Thymus vulgaris | | Descriptor: | 1,2-ETHANEDIOL, Putative gamma-terpinene synthase | | Authors: | Parthier, C.P, Rudolph, K, Muller, Y.A, Mueller-Uri, F. | | Deposit date: | 2015-06-12 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Expression, crystallization and structure elucidation of gamma-terpinene synthase from Thymus vulgaris.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6BBS
 
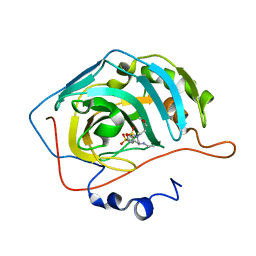 | | Joint X-ray/neutron structure of human carbonic anhydrase II in complex with brinzolamide | | Descriptor: | (+)-4-ETHYLAMINO-3,4-DIHYDRO-2-(METHOXY)PROPYL-2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Kovalevsky, A, Aggarwal, M, McKenna, R. | | Deposit date: | 2017-10-19 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | "To Be or Not to Be" Protonated: Atomic Details of Human Carbonic Anhydrase-Clinical Drug Complexes by Neutron Crystallography and Simulation.
Structure, 26, 2018
|
|
6BBZ
 
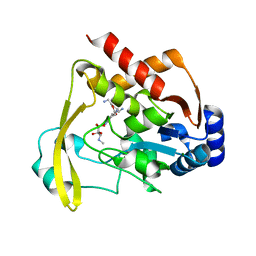 | | Room temperature neutron/X-ray structure of sisomicin bound AAC-VIa | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, AAC 3-VI protein, MAGNESIUM ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2017-10-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | A low-barrier hydrogen bond mediates antibiotic resistance in a noncanonical catalytic triad.
Sci Adv, 4, 2018
|
|
6BC4
 
 | |
6APZ
 
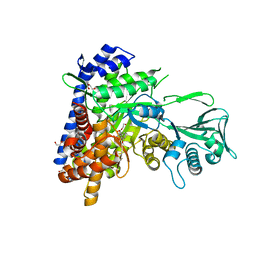 | |
5F3F
 
 | | Crystal structure of para-biphenyl-2-methyl-3'-methyl amide mannoside bound to FimH lectin domain | | Descriptor: | 3-[4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3-methyl-phenyl]-~{N}-methyl-benzamide, Protein FimH | | Authors: | Klein, R.D, Hultgren, S.J. | | Deposit date: | 2015-12-02 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Antivirulence C-Mannosides as Antibiotic-Sparing, Oral Therapeutics for Urinary Tract Infections.
J.Med.Chem., 59, 2016
|
|
6AXP
 
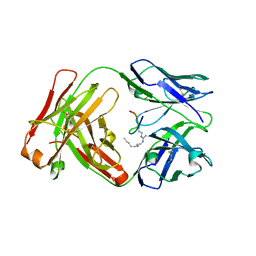 | |
5CBP
 
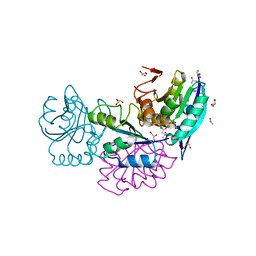 | | Crystal Structure of Conjoint Pyrococcus furiosus L-asparaginase at 37 degree C | | Descriptor: | CITRATE ANION, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Sharma, P, Yadav, S.P, Tomar, R, Kundu, B, Ashish, F. | | Deposit date: | 2015-07-01 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.358 Å) | | Cite: | Heat induces end to end repetitive association in P. furiosus L-asparaginase which enables its thermophilic property.
Sci Rep, 10, 2020
|
|
4XBS
 
 | | 2-deoxyribose-5-phosphate aldolase mutant - E78K | | Descriptor: | Deoxyribose-phosphate aldolase | | Authors: | Jiao, X.-C, Pan, J, Xu, G.-C, Kong, X.-D, Chen, Q, Zhang, Z.-J, Xu, J.-H. | | Deposit date: | 2014-12-17 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Efficient synthesis of a statin precursor in high space-time yield by a new aldehyde-tolerant aldolase identified from Lactobacillus brevis
Catalysis Science And Technology, 5, 2015
|
|
4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
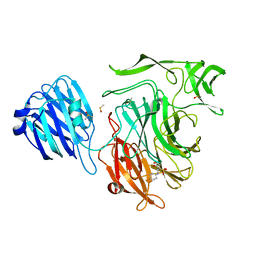
4XE9
 
 | |
5CJF
 
 | | The crystal structure of the human carbonic anhydrase XIV in complex with a 1,1'-biphenyl-4-sulfonamide inhibitor. | | Descriptor: | 4'-(4-aminobenzoyl)biphenyl-4-sulfonamide, Carbonic anhydrase 14, GLYCEROL, ... | | Authors: | Alterio, V, De Simone, G. | | Deposit date: | 2015-07-14 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of 1,1'-Biphenyl-4-sulfonamides as a New Class of Potent and Selective Carbonic Anhydrase XIV Inhibitors.
J.Med.Chem., 58, 2015
|
|
6BDP
 
 | | Schistosoma mansoni (Blood Fluke) Sulfotransferase/CIDD-0000071 (Compound 9c) Complex | | Descriptor: | (4-{[(3R)-1-benzylpyrrolidin-3-yl]amino}-2-nitrophenyl)methanol, ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase oxamniquine resistance protein | | Authors: | Taylor, A.B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Design, Synthesis, and Characterization of Novel Small Molecules as Broad Range Antischistosomal Agents.
ACS Med Chem Lett, 9, 2018
|
|
7B0B
 
 | | Fab HbnC3t1p1_C6 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, IGK@ protein, ... | | Authors: | Borenstein-Katz, A, Diskin, R. | | Deposit date: | 2020-11-19 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Somatic hypermutation introduces bystander mutations that prepare SARS-CoV-2 antibodies for emerging variants.
Immunity, 56, 2023
|
|
4XSM
 
 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with D-talitol | | Descriptor: | D-altritol, D-tagatose 3-epimerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
6B67
 
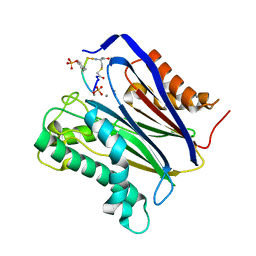 | | Human PP2Calpha (PPM1A) complexed with cyclic peptide c(MpSIpYVA) | | Descriptor: | CALCIUM ION, Protein phosphatase 1A, cyclic peptide c(MpSIpYVA) | | Authors: | Dyda, F, Kosek, D. | | Deposit date: | 2017-10-01 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A trapped human PPM1A-phosphopeptide complex reveals structural features critical for regulation of PPM protein phosphatase activity.
J. Biol. Chem., 293, 2018
|
|
6APN
 
 | |
2DKD
 
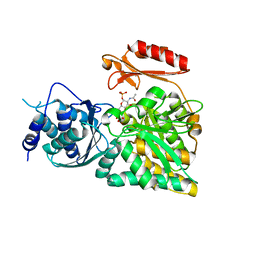 | | Crystal structure of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, in the product complex | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-galactopyranose, PHOSPHATE ION, Phosphoacetylglucosamine mutase, ... | | Authors: | Nishitani, Y, Maruyama, D, Nonaka, T, Kita, A, Fukami, T.A, Mio, T, Yamada-Okabe, H, Yamada-Okabe, T, Miki, K. | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of N-Acetylglucosamine-phosphate Mutase, a Member of the {alpha}-D-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes.
J.Biol.Chem., 281, 2006
|
|
6AUG
 
 | |
6AVA
 
 | |
6AW0
 
 | | Crystal structure of CEACAM3 L44Q | | Descriptor: | CHLORIDE ION, Carcinoembryonic antigen-related cell adhesion molecule 3, GLYCEROL, ... | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
5FEF
 
 | |
5FDS
 
 | |
6AY7
 
 | | Cartilage homing cysteine-dense-peptides | | Descriptor: | CHLORIDE ION, Potassium channel toxin alpha-KTx 3.5, SULFATE ION, ... | | Authors: | Gewe, M.M, Strong, R.K. | | Deposit date: | 2017-09-07 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Cartilage homing cysteine-dense-peptides
To Be Published
|
|
4XHB
 
 | |
