6FJT
 
 | | 4-chloro-benzamidine in complex with thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-chloranylbenzenecarboximidamide, DIMETHYL SULFOXIDE, ... | | Authors: | Abazi, N, Heine, A, Klebe, G. | | Deposit date: | 2018-01-23 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | 4-chloro-benzamidine in complex with thrombin
To be published
|
|
6FAK
 
 | | Human afamin orthorhombic crystal form by controlled hydration | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rupp, B, Naschberger, A, Bowler, M.W. | | Deposit date: | 2017-12-15 | | Release date: | 2019-01-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Controlled dehydration, structural flexibility and gadolinium MRI contrast compound binding in the human plasma glycoprotein afamin.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6ZUM
 
 | | Crystal structure of dimethylated RSL-N23H (RSL-B3) in complex with cucurbit[7]uril and zinc | | Descriptor: | ACETATE ION, Fucose-binding lectin protein, SODIUM ION, ... | | Authors: | Guagnini, F, Engilberge, S, Flood, R.J, Ramberg, K.O, Crowley, P.B. | | Deposit date: | 2020-07-23 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Metal-Mediated Protein-Cucurbituril Crystalline Architectures
Cryst.Growth Des., 2020
|
|
6ZY0
 
 | | Catabolic reductive dehalogenase NpRdhA, N-terminally tagged, K488Q variant | | Descriptor: | CHLORIDE ION, COBALAMIN, IRON/SULFUR CLUSTER, ... | | Authors: | Leys, D, Halliwell, T. | | Deposit date: | 2020-07-30 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Catabolic Reductive Dehalogenase Substrate Complex Structures Underpin Rational Repurposing of Substrate Scope.
Microorganisms, 8, 2020
|
|
7BJ3
 
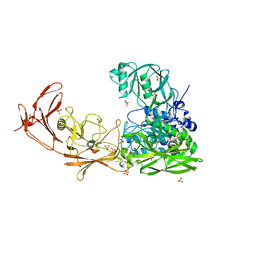 | | ScpA from Streptococcus pyogenes, S512A active site mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, C5a peptidase, CALCIUM ION, ... | | Authors: | Kagawa, T.F, O'Connell, M.R, Cooney, J.C. | | Deposit date: | 2021-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Enzyme kinetic and binding studies identify determinants of specificity for the immunomodulatory enzyme ScpA, a C5a inactivating bacterial protease.
Comput Struct Biotechnol J, 19, 2021
|
|
7A1B
 
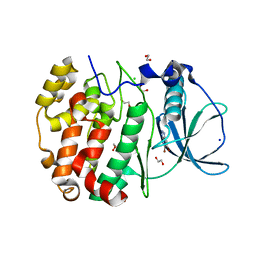 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the ATP-competitive inhibitor 5,6-dibromo-1H-triazolo[4,5-b]pyridine | | Descriptor: | 1,2-ETHANEDIOL, 5,6-dibromo-1H-triazolo[4,5-b]pyridine, CHLORIDE ION, ... | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-12 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.287 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
7A70
 
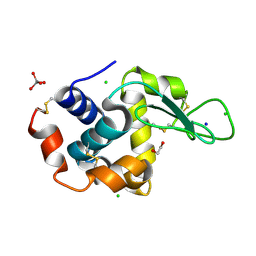 | | HEW lysozyme in complex with Ti(OH)4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme, ... | | Authors: | Calderone, V, Gigli, L, Ravera, E, Luchinat, C. | | Deposit date: | 2020-08-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | On the Mechanism of Bioinspired Formation of Inorganic Oxides: Structural Evidence of the Electrostatic Nature of the Interaction between a Mononuclear Inorganic Precursor and Lysozyme.
Biomolecules, 11, 2020
|
|
7A76
 
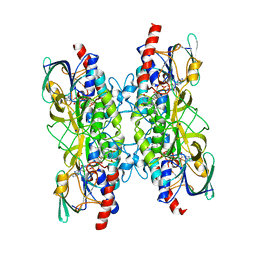 | |
6K2O
 
 | | Structural basis of glycan recognition in globally predominant human P[8] rotavirus | | Descriptor: | Outer capsid protein VP4, SODIUM ION, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose | | Authors: | Duan, Z, Sun, X. | | Deposit date: | 2019-05-15 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Structural Basis of Glycan Recognition in Globally Predominant Human P[8] Rotavirus.
Virol Sin, 35, 2020
|
|
6ZRX
 
 | | Crystal structure of 6-dimethylallyltryptophan synthase from Micromonospora olivasterospora in complex with DMASPP and Trp | | Descriptor: | DI(HYDROXYETHYL)ETHER, DMATS type aromatic prenyltransferase, S-(3-methylbut-2-en-1-yl) trihydrogen thiodiphosphate, ... | | Authors: | Ostertag, E, Stehle, T, Zocher, G. | | Deposit date: | 2020-07-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reprogramming Substrate and Catalytic Promiscuity of Tryptophan Prenyltransferases.
J.Mol.Biol., 433, 2020
|
|
7BDR
 
 | | Structure of CTX-M-15 E166Q mutant crystallised in the presence of tazobactam (AAI101) | | Descriptor: | Beta-lactamase, CHLORIDE ION, SODIUM ION, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-12-22 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Penicillanic Acid Sulfones Inactivate the Extended-Spectrum beta-Lactamase CTX-M-15 through Formation of a Serine-Lysine Cross-Link: an Alternative Mechanism of beta-Lactamase Inhibition.
Mbio, 13, 2022
|
|
7BIZ
 
 | |
7BM0
 
 | | Crystal structure of the tick-borne encephalitis virus NS3 helicase in complex with AMPPNP | | Descriptor: | MANGANESE (II) ION, NS3 helicase domain, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Anindita, P.D, Grinkevich, P, Franta, Z. | | Deposit date: | 2021-01-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insight into the RNA-stimulated ATPase activity of tick-borne encephalitis virus helicase.
J.Biol.Chem., 298, 2022
|
|
7C67
 
 | | Crystal structure of beta-glycosides-binding protein of ABC transporter in a closed state bound to cellotriose | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
7AQ4
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583E | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
6FWQ
 
 | | Structure of an E333Q variant of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-1,3-mannobiose and alpha-1,2-mannobiose | | Descriptor: | Glycosyl hydrolase family 71, SODIUM ION, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose, ... | | Authors: | Sobala, L.F, Speciale, G, Hakki, Z, Fernandes, P.Z, Raich, L, Rojas-Cervellera, V, Bennet, A, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Lu, D, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2018-03-07 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An Epoxide Intermediate in Glycosidase Catalysis.
Acs Cent.Sci., 6, 2020
|
|
7AQ3
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583D | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
6GBW
 
 | | Thrombin in complex with MI2100 ((S)-N-(2-(aminomethyl)-5-chlorobenzyl)-1-((benzylsulfonyl)-L-arginyl)pyrrolidine-2-carboxamide) | | Descriptor: | (2~{S})-~{N}-[[2-(aminomethyl)-5-chloranyl-phenyl]methyl]-1-[(2~{S})-5-carbamimidamido-2-[(phenylmethyl)sulfonylamino]pentanoyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G. | | Deposit date: | 2018-04-16 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Strategies for Late-Stage Optimization: Profiling Thermodynamics by Preorganization and Salt Bridge Shielding.
J.Med.Chem., 62, 2019
|
|
7AEW
 
 | |
7APY
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, D576A | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.778 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
7AQA
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H382A | | Descriptor: | (dicuprio-$l^{3}-sulfanyl)-sulfanyl-copper, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
7B3U
 
 | | OXA-10 beta-lactamase with covalent modification | | Descriptor: | Beta-lactamase OXA-10, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Lang, P.A, Brem, J, Schofield, C.J. | | Deposit date: | 2020-12-01 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Studies on enmetazobactam clarify mechanisms of widely used beta-lactamase inhibitors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7B3S
 
 | | OXA-10 beta-lactamase with S67Dha modification | | Descriptor: | Beta-lactamase OXA-10, CARBON DIOXIDE, SODIUM ION, ... | | Authors: | Lang, P.A, Brem, J, Schofield, C.J. | | Deposit date: | 2020-12-01 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Studies on enmetazobactam clarify mechanisms of widely used beta-lactamase inhibitors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7B3R
 
 | |
7B7H
 
 | | The glucuronoyl esterase OtCE15A R268A variant from Opitutus terrae in complex with, and covalently linked to, D-glucuronate | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, GLYCINE, ... | | Authors: | Mazurkewich, S, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2020-12-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism and biomass association of glucuronoyl esterase: an alpha / beta hydrolase with potential in biomass conversion.
Nat Commun, 13, 2022
|
|
