1JD4
 
 | | Crystal Structure of DIAP1-BIR2 | | Descriptor: | APOPTOSIS 1 INHIBITOR, ZINC ION | | Authors: | Wu, J.W, Cocina, A.E, Chai, J, Hay, B.A, Shi, Y. | | Deposit date: | 2001-06-12 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides.
Mol.Cell, 8, 2001
|
|
1JD5
 
 | | Crystal Structure of DIAP1-BIR2/GRIM | | Descriptor: | APOPTOSIS 1 INHIBITOR, ZINC ION, cell death protein GRIM | | Authors: | Wu, J.W, Cocina, A.E, Chai, J, Hay, B.A, Shi, Y. | | Deposit date: | 2001-06-12 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides.
Mol.Cell, 8, 2001
|
|
1JD6
 
 | | Crystal Structure of DIAP1-BIR2/Hid Complex | | Descriptor: | APOPTOSIS 1 INHIBITOR, ZINC ION, head involution defective protein | | Authors: | Wu, J.W, Cocina, A.E, Chai, J, Hay, B.A, Shi, Y. | | Deposit date: | 2001-06-12 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides.
Mol.Cell, 8, 2001
|
|
1JD7
 
 | |
1JD8
 
 | | Solution structure of lactam analogue DapD of HIV gp41 600-612 loop | | Descriptor: | Transmembrane protein gp41 | | Authors: | Phan Chan Du, A, Limal, D, Semetey, V, Dali, H, Jolivet, M, Desgranges, C, Cung, M.T, Briand, J.P, Petit, M.C, Muller, S. | | Deposit date: | 2001-06-13 | | Release date: | 2003-07-01 | | Last modified: | 2018-10-10 | | Method: | SOLUTION NMR | | Cite: | Structural and immunological characterisation of heteroclitic peptide analogues corresponding to the 600-612 region of the HIV envelope gp41 glycoprotein.
J.Mol.Biol., 323, 2002
|
|
1JD9
 
 | |
1JDA
 
 | | MALTOTETRAOSE-FORMING EXO-AMYLASE | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
1JDB
 
 | | CARBAMOYL PHOSPHATE SYNTHETASE FROM ESCHERICHIA COLI | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMOYL PHOSPHATE SYNTHETASE, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Holden, H.M, Wesenberg, G, Raushel, F.M, Rayment, I. | | Deposit date: | 1997-03-25 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of carbamoyl phosphate synthetase determined to 2.1 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1JDC
 
 | | MUTANT (E219Q) MALTOTETRAOSE-FORMING EXO-AMYLASE COCRYSTALLIZED WITH MALTOTETRAOSE (CRYSTAL TYPE 1) | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
1JDD
 
 | | MUTANT (E219Q) MALTOTETRAOSE-FORMING EXO-AMYLASE COCRYSTALLIZED WITH MALTOTETRAOSE (CRYSTAL TYPE 2) | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
1JDE
 
 | | K22A mutant of pyruvate, phosphate dikinase | | Descriptor: | PYRUVATE, PHOSPHATE DIKINASE, SULFATE ION | | Authors: | Ye, D, Wei, M, McGuire, M, Huang, K, Kapadia, G, Herzberg, O, Martin, B.M, Dunaway-Mariano, D. | | Deposit date: | 2001-06-13 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Investigation of the catalytic site within the ATP-grasp domain of Clostridium symbiosum pyruvate phosphate dikinase.
J.Biol.Chem., 276, 2001
|
|
1JDF
 
 | | Glucarate Dehydratase from E.coli N341D mutant | | Descriptor: | 2,3-DIHYDROXY-5-OXO-HEXANEDIOATE, Glucarate Dehydratase, ISOPROPYL ALCOHOL, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2001-06-13 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in the active site of D-glucarate dehydratase from Escherichia coli.
Biochemistry, 40, 2001
|
|
1JDG
 
 | | Solution Structure of a Trans-Opened (10S)-dA Adduct of (+)-(7S,8R,9S,10R)-7,8-Dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene in a fully Complementary DNA Duplex | | Descriptor: | 5'-D(*CP*CP*TP*CP*GP*TP*GP*AP*CP*CP*G)-3', 5'-D(*CP*GP*GP*TP*CP*(BPA)AP*CP*GP*AP*GP*G)-3', 7S,8R,9R-TRIHYDROXY-7,8,9,10-TETRAHYDRO BENZO[A]PYRENE | | Authors: | Pradhan, P, Tirumala, S, Liu, X, Sayer, J.M, Jerina, D.M, Yeh, H.J.C. | | Deposit date: | 2001-06-13 | | Release date: | 2001-07-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a trans-opened (10S)-dA adduct of (+)-(7S,8R,9S,10R)-7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene in a fully complementary DNA duplex: evidence for a major syn conformation.
Biochemistry, 40, 2001
|
|
1JDH
 
 | | CRYSTAL STRUCTURE OF BETA-CATENIN AND HTCF-4 | | Descriptor: | BETA-CATENIN, hTcf-4 | | Authors: | Graham, T.A, Ferkey, D.M, Mao, F, Kimelman, D, Xu, W. | | Deposit date: | 2001-06-13 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tcf4 can specifically recognize beta-catenin using alternative conformations.
Nat.Struct.Biol., 8, 2001
|
|
1JDI
 
 | | CRYSTAL STRUCTURE OF L-RIBULOSE-5-PHOSPHATE 4-EPIMERASE | | Descriptor: | L-RIBULOSE 5 PHOSPHATE 4-EPIMERASE, ZINC ION | | Authors: | Luo, Y, Samuel, J, Mosimann, S.C, Lee, J.E, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 2001-06-13 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of L-ribulose-5-phosphate 4-epimerase: an aldolase-like platform for epimerization.
Biochemistry, 40, 2001
|
|
1JDJ
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MEXICANA GLYCEROL-3-PHOSPHATE DEHYDROGENASE IN COMPLEX WITH 2-FLUORO-6-CHLOROPURINE | | Descriptor: | 6-CHLORO-2-FLUOROPURINE, GLYCEROL-3-PHOSPHATE DEHYDROGENASE, PENTADECANE | | Authors: | Suresh, S, Wisedchaisri, G, Kennedy, K.J, Verlinde, C.L.M.J, Gelb, M.H, Hol, W.G.J. | | Deposit date: | 2001-06-14 | | Release date: | 2002-06-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anomalous differences of light elements in determining precise binding modes of ligands to glycerol-3-phosphate dehydrogenase.
Chem.Biol., 9, 2002
|
|
1JDK
 
 | | solution structure of lactam analogue (EDap) of HIV gp41 600-612 loop. | | Descriptor: | ACETYL GROUP | | Authors: | Phan Chan Du, A, Limal, D, Semetey, V, Dali, H, Jolivet, M, Desgranges, C, Cung, M.T, Briand, J.P, Petit, M.C, Muller, S. | | Deposit date: | 2001-06-14 | | Release date: | 2003-07-01 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural and immunological characterisation of heteroclitic peptide analogues corresponding to the 600-612 region of the HIV envelope gp41 glycoprotein.
J.Mol.Biol., 323, 2002
|
|
1JDL
 
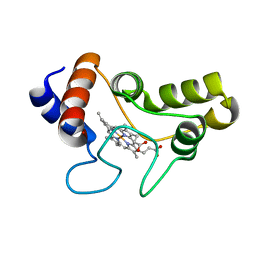 | | Structure of cytochrome c2 from Rhodospirillum Centenum | | Descriptor: | CYTOCHROME C2, ISO-2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Camara-Artigas, A, Williams, J.C, Allen, J.P. | | Deposit date: | 2001-06-14 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of cytochrome c2 from Rhodospirillum centenum.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1JDM
 
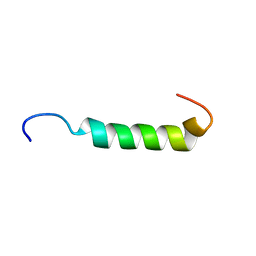 | | NMR Structure of Sarcolipin | | Descriptor: | Sarcolipin | | Authors: | Veglia, G, Mascioni, A. | | Deposit date: | 2001-06-14 | | Release date: | 2002-02-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and orientation of sarcolipin in lipid environments.
Biochemistry, 41, 2002
|
|
1JDN
 
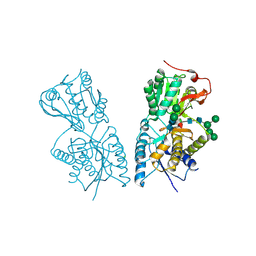 | | Crystal Structure of Hormone Receptor | | Descriptor: | ATRIAL NATRIURETIC PEPTIDE CLEARANCE RECEPTOR, CHLORIDE ION, beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | He, X.-L, Chow, D.-C, Martick, M.M, Garcia, K.C. | | Deposit date: | 2001-06-14 | | Release date: | 2001-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric activation of a spring-loaded natriuretic peptide receptor dimer by hormone.
Science, 293, 2001
|
|
1JDO
 
 | |
1JDP
 
 | | Crystal Structure of Hormone/Receptor Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATRIAL NATRIURETIC PEPTIDE CLEARANCE RECEPTOR, ... | | Authors: | He, X.-L, Chow, D.-C, Martick, M.M, Garcia, K.C. | | Deposit date: | 2001-06-14 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric activation of a spring-loaded natriuretic peptide receptor dimer by hormone.
Science, 293, 2001
|
|
1JDQ
 
 | | Solution Structure of TM006 Protein from Thermotoga maritima | | Descriptor: | HYPOTHETICAL PROTEIN TM0983 | | Authors: | Denisov, A.Y, Finak, G, Yee, A, Kozlov, G, Gehring, K, Arrowsmith, C.H. | | Deposit date: | 2001-06-14 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JDR
 
 | | Crystal Structure of a Proximal Domain Potassium Binding Variant of Cytochrome c Peroxidase | | Descriptor: | Cytochrome c Peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bonagura, C.A, Sundaramoorthy, M, Bhaskar, B, Poulos, T.L. | | Deposit date: | 2001-06-14 | | Release date: | 2001-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The effects of an engineered cation site on the structure, activity, and EPR properties of cytochrome c peroxidase.
Biochemistry, 38, 1999
|
|
1JDS
 
 | | 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE COMPLEX WITH PHOSPHATE (SPACE GROUP P21) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-METHYLTHIOADENOSINE PHOSPHORYLASE, PHOSPHATE ION | | Authors: | Appleby, T.C, Mathews, I.I, Porcelli, M, Cacciapuoti, G, Ealick, S.E. | | Deposit date: | 2001-06-15 | | Release date: | 2001-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of a hyperthermophilic 5'-deoxy-5'-methylthioadenosine phosphorylase from Sulfolobus solfataricus.
J.Biol.Chem., 276, 2001
|
|
