5NFP
 
 | | Glucocorticoid Receptor in complex with budesonide | | Descriptor: | (1~{S},2~{S},4~{R},6~{R},8~{S},9~{S},11~{S},12~{S},13~{R})-9,13-dimethyl-11-oxidanyl-8-(2-oxidanylethanoyl)-6-propyl-5,7-dioxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icosa-14,17-dien-16-one, 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, ... | | Authors: | Edman, K, Wissler, L. | | Deposit date: | 2017-03-15 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective Nonsteroidal Glucocorticoid Receptor Modulators for the Inhaled Treatment of Pulmonary Diseases.
J. Med. Chem., 60, 2017
|
|
2WDF
 
 | | Termus thermophilus Sulfate thiohydrolase SoxB | | Descriptor: | MANGANESE (II) ION, SULFUR OXIDATION PROTEIN SOXB, TERTIARY-BUTYL ALCOHOL | | Authors: | Sauve, V, Roversi, P, Leath, K.J, Garman, E.F, Antrobus, R, Lea, S.M, Berks, B.C. | | Deposit date: | 2009-03-24 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Mechanism for the Hydrolysis of a Sulfur-Sulfur Bond Based on the Crystal Structure of the Thiosulfohydrolase Soxb.
J.Biol.Chem., 284, 2009
|
|
1XYU
 
 | | Solution structure of the sheep prion protein with polymorphism H168 | | Descriptor: | Major prion protein | | Authors: | Calzolai, L, Lysek, D.A, Guntert, P, Wuthrich, K. | | Deposit date: | 2004-11-11 | | Release date: | 2005-01-04 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of cats, dogs, pigs, and sheep
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
4IVH
 
 | | Crystal structure of QKLVFFAED nonapeptide segment from amyloid beta incorporated into a macrocyclic beta-sheet template | | Descriptor: | SODIUM ION, TERTIARY-BUTYL ALCOHOL, cyclo[Gln-Lys-Leu-Val-Phe-Phe-Ala-Glu-Asp-(delta-linked-Orn)-Hao-Lys-Hao-(p-bromoPhe)-Thr-(delta-linked-Orn)] | | Authors: | Pham, J.D, Chim, N, Goulding, C.W, Nowick, J.S. | | Deposit date: | 2013-01-22 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structures of beta-Amyloid Peptide Oligomers
J.Am.Chem.Soc., 2013
|
|
3GST
 
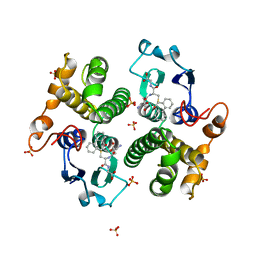 | | STRUCTURE OF THE XENOBIOTIC SUBSTRATE BINDING SITE OF A GLUTATHIONE S-TRANSFERASE AS REVEALED BY X-RAY CRYSTALLOGRAPHIC ANALYSIS OF PRODUCT COMPLEXES WITH THE DIASTEREOMERS OF 9-(S-GLUTATHIONYL)-10-HYDROXY-9, 10-DIHYDROPHENANTHRENE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, GLUTATHIONE S-TRANSFERASE, SULFATE ION | | Authors: | Ji, X, Ammon, H.L, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1993-06-07 | | Release date: | 1993-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the xenobiotic substrate binding site of a glutathione S-transferase as revealed by X-ray crystallographic analysis of product complexes with the diastereomers of 9-(S-glutathionyl)-10-hydroxy-9,10-dihydrophenanthrene.
Biochemistry, 33, 1994
|
|
3X1J
 
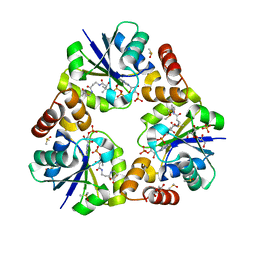 | |
1Y9A
 
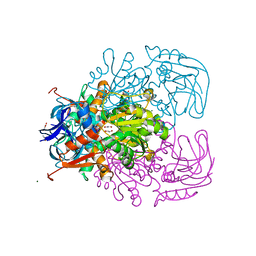 | | Alcohol Dehydrogenase from Entamoeba histolotica in complex with cacodylate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CACODYLATE ION, ... | | Authors: | Shimon, L.J, Peretz, M, Goihberg, E, Burstein, Y, Frolow, F. | | Deposit date: | 2004-12-15 | | Release date: | 2006-01-17 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of alcohol dehydrogenase from Entamoeba histolytica.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4JJC
 
 | |
1Y2S
 
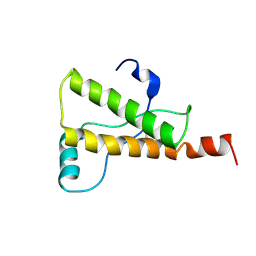 | | Ovine Prion Protein Variant R168 | | Descriptor: | Major prion protein | | Authors: | Christen, B, Lysek, D.A, Herrmann, T, Wuthrich, K. | | Deposit date: | 2004-11-23 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of cats, dogs, pigs, and sheep
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1HMH
 
 | |
3PB0
 
 | |
4E3T
 
 | |
1H89
 
 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX2 | | Descriptor: | CAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), DNA(5'-(*GP*AP*TP*GP*TP*GP*GP*CP*GP*CP*AP* AP*TP*CP*CP*TP*TP*AP*AP*CP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2001-01-30 | | Release date: | 2002-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
4MJO
 
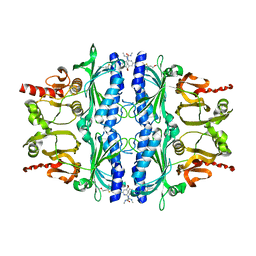 | | Human liver fructose-1,6-bisphosphatase(d-fructose-1,6-bisphosphate, 1-phosphohydrolase) (e.c.3.1.3.11) complexed with the allosteric inhibitor 3 | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-({4-bromo-6-[(methylcarbamoyl)amino]pyridin-2-yl}carbamoyl)-5-(2-methoxyethyl)-4-methylthiophene-2-sulfonamide | | Authors: | Ruf, A, Joseph, C, Tetaz, T, Benz, J. | | Deposit date: | 2013-09-04 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determination of protein-ligand binding constants of a cooperatively regulated tetrameric enzyme using electrospray mass spectrometry.
Acs Chem.Biol., 9, 2014
|
|
4MM1
 
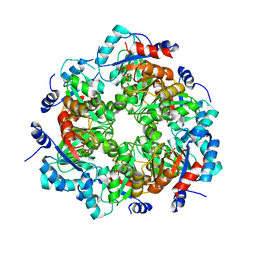 | | GGGPS from Methanothermobacter thermautotrophicus | | Descriptor: | Geranylgeranylglyceryl phosphate synthase, SN-GLYCEROL-1-PHOSPHATE, TRIETHYLENE GLYCOL | | Authors: | Rajendran, C, Peterhoff, D, Beer, B, Kumpula, E.P, Kapetaniou, E, Guldan, H, Wierenga, R.K, Sterner, R, Babinger, P. | | Deposit date: | 2013-09-07 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8004 Å) | | Cite: | A comprehensive analysis of the geranylgeranylglyceryl phosphate synthase enzyme family identifies novel members and reveals mechanisms of substrate specificity and quaternary structure organization.
Mol.Microbiol., 92, 2014
|
|
5OIP
 
 | |
6ZNR
 
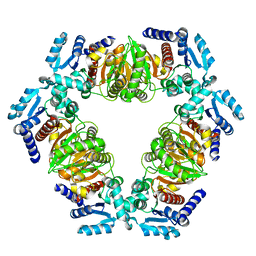 | | MaeB PTA domain R535A mutant | | Descriptor: | Malate dehydrogenase | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.217 Å) | | Cite: | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
6ZNJ
 
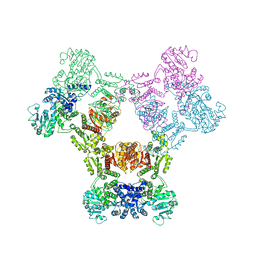 | | MaeB full-length enzyme apoprotein form | | Descriptor: | NADP-dependent malate dehydrogenase,Malate dehydrogenase | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
1EE2
 
 | |
6ZN7
 
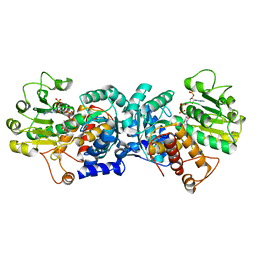 | | MaeB malic enzyme domain apoprotein | | Descriptor: | MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent malate dehydrogenase,Malate dehydrogenase | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
6ZN4
 
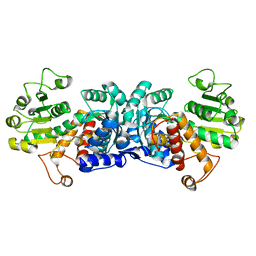 | | MaeB malic enzyme domain apoprotein | | Descriptor: | MAGNESIUM ION, NADP-dependent malate dehydrogenase,Malate dehydrogenase | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
6ROB
 
 | | Human Carbonic Anhydrase II in complex with 4-cyanobenzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 4-cyanobenzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-05-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.929 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
6ZN9
 
 | | MaeB PTA domain apoprotein | | Descriptor: | Malate dehydrogenase | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
6ZNU
 
 | | MaeB PTA domain E544R mutant | | Descriptor: | ACETYL COENZYME *A, CHLORIDE ION, Malate dehydrogenase | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
6ZNG
 
 | | MaeB full-length acetyl-CoA bound state | | Descriptor: | ACETYL COENZYME *A, NADP-dependent malate dehydrogenase,Malate dehydrogenase | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
