3ATT
 
 | | Crystal structure of Rv3168 with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Kim, Y.-G, Kim, S, Nguyen, C.M.T, Kim, K.-J. | | Deposit date: | 2011-01-13 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis Rv3168: a putative aminoglycoside antibiotics resistance enzyme
Proteins, 79, 2011
|
|
2ZUV
 
 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase in complex with GlcNAc, Ethylene glycol, and nitrate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, Lacto-N-biose phosphorylase, ... | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
4O13
 
 | | The crystal structure of NAMPT in complex with GNE-618 | | Descriptor: | 1,2-ETHANEDIOL, N-(4-{[3-(trifluoromethyl)phenyl]sulfonyl}benzyl)-2H-pyrazolo[3,4-b]pyridine-5-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O3N
 
 | | Crystal structure of human dna polymerase eta in ternary complex with native dna and incoming nucleotide (dcp) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*GP*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | Kinetics, Structure, and Mechanism of 8-Oxo-7,8-dihydro-2'-deoxyguanosine Bypass by Human DNA Polymerase eta
J.Biol.Chem., 289, 2014
|
|
2ZUU
 
 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, GLYCEROL, Lacto-N-biose phosphorylase, ... | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
4O09
 
 | | Identification of novel HSP90 / isoform selective inhibitors using structure-based drug design. Demonstration of potential utility in treating CNS disorders such as Huntington s disease | | Descriptor: | 8-(2-methylpropyl)-6-(3,6,6-trimethyl-4-oxo-4,5,6,7-tetrahydro-1H-indol-1-yl)-3,4-dihydroisoquinolin-1(2H)-one, Heat shock protein HSP 90-alpha | | Authors: | Zuccola, H.J, Ernst, J.T. | | Deposit date: | 2013-12-13 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identification of Novel HSP90 alpha / beta Isoform Selective Inhibitors Using Structure-Based Drug Design. Demonstration of Potential Utility in Treating CNS Disorders such as Huntington's Disease.
J.Med.Chem., 57, 2014
|
|
4O3R
 
 | | Crystal structure of human polymerase eta extending an 8-oxog dna lesion: post insertion of 8-oxog-da pair | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*A)-3'), DNA (5'-D(*CP*AP*TP*GP*(8OG)P*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Kinetics, Structure, and Mechanism of 8-Oxo-7,8-dihydro-2'-deoxyguanosine Bypass by Human DNA Polymerase eta
J.Biol.Chem., 289, 2014
|
|
2Z7L
 
 | |
2ZEQ
 
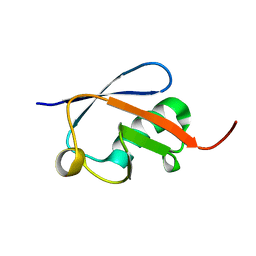 | | Crystal structure of ubiquitin-like domain of murine Parkin | | Descriptor: | E3 ubiquitin-protein ligase parkin | | Authors: | Tomoo, K. | | Deposit date: | 2007-12-14 | | Release date: | 2008-08-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure and molecular dynamics simulation of ubiquitin-like domain of murine parkin
Biochim.Biophys.Acta, 1784, 2008
|
|
4NP9
 
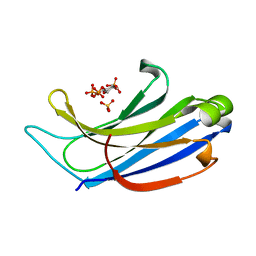 | | Structure of Rabphilin C2A domain bound to IP3 | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Rabphilin-3A, SULFATE ION | | Authors: | Guillen, J, Ferrer-Orta, C, Buxaderas, M, Perez-Sanchez, D, Guerrero-Valero, M, Luengo-Gil, G, Pous, J, Guerra, P, Gomez-Fernandez, J.C, Verdaguer, N, Corbalan-Garcia, S. | | Deposit date: | 2013-11-21 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural insights into the Ca2+ and PI(4,5)P2 binding modes of the C2 domains of rabphilin 3A and synaptotagmin 1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2ZNU
 
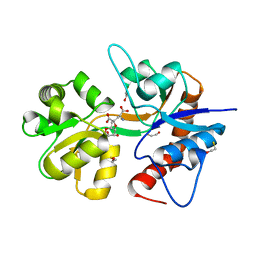 | | Crystal structure of the ligand-binding core of the human ionotropic glutamate receptor, GluR5, in complex with a novel selective agonist, neodysiherbaine A | | Descriptor: | (2R,3aR,6R,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6,7-dihydroxyhexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, BETA-MERCAPTOETHANOL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2008-05-01 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
4O10
 
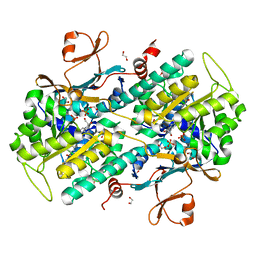 | | Structural and Biochemical Analyses of the Catalysis and Potency Impact of Inhibitor Phosphoribosylation by Human Nicotinamide Phosphoribosyltransferase | | Descriptor: | 1,2-ETHANEDIOL, N-{4-[(3,5-difluorophenyl)sulfonyl]benzyl}indolizine-7-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Wang, W. | | Deposit date: | 2013-12-14 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and biochemical analyses of the catalysis and potency impact of inhibitor phosphoribosylation by human nicotinamide phosphoribosyltransferase.
Chembiochem, 15, 2014
|
|
2ZT1
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 100 K: Laser on [810 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4NZK
 
 | |
2ZNT
 
 | | Crystal structure of the ligand-binding core of the human ionotropic glutamate receptor, GluR5, in complex with a novel selective agonist, dysiherbaine | | Descriptor: | (2R,3aR,6S,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6-hydroxy-7-(methylamino)hexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, BETA-MERCAPTOETHANOL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2008-05-01 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
4O17
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O28
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-{4-[(3,5-difluorophenyl)sulfonyl]benzyl}imidazo[1,2-a]pyridine-6-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-17 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O38
 
 | | Crystal structure of the human cyclin G associated kinase (GAK) | | Descriptor: | Cyclin-G-associated kinase, GLYCEROL, SUCCINIC ACID | | Authors: | Zhang, R, Hatzos-Skintges, C, Weger, A, Chaikuad, A, Eswaran, J, Fedorov, O, King, O, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-18 | | Release date: | 2014-01-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of the human cyclin G associated kinase (GAK)
To be Published
|
|
4NV1
 
 | | Crystal structure of a 4-N formyltransferase from Francisella tularensis | | Descriptor: | Formyltransferase, PHOSPHATE ION, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Zimmer, A.L, Holden, H.M. | | Deposit date: | 2013-12-04 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of a sugar N-formyltransferase from Francisella tularensis.
Protein Sci., 23, 2014
|
|
3AS0
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - W275G mutant complex structure with Sanguinarine | | Descriptor: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3AVA
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-02 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AVI
 
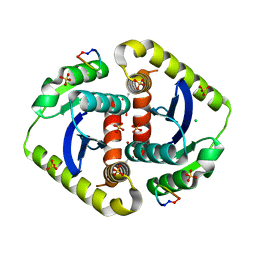 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
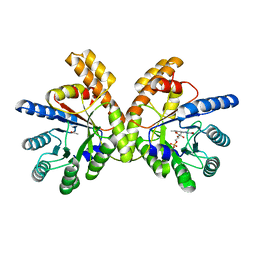
3APT
 
 | |
3APG
 
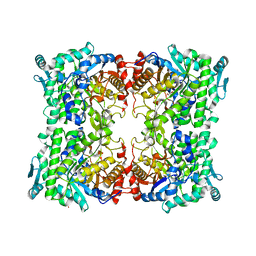 | |
3ARS
 
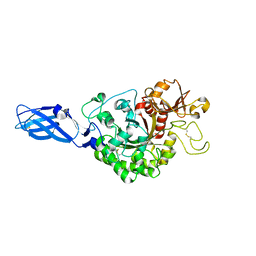 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - apo structure of mutant W275G | | Descriptor: | Chitinase A | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
