6GEB
 
 | |
2D28
 
 | | Structure of the N-terminal domain of XpsE (crystal form P43212) | | Descriptor: | CACODYLATE ION, type II secretion ATPase XpsE | | Authors: | Chen, Y, Shiue, S.-J, Huang, C.-W, Chang, J.-L, Chien, Y.-L, Hu, N.-T, Chan, N.-L. | | Deposit date: | 2005-09-03 | | Release date: | 2005-09-20 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of the XpsE N-Terminal Domain, an Essential Component of the Xanthomonas campestris Type II Secretion System
J.Biol.Chem., 280, 2005
|
|
2BCM
 
 | | DaaE adhesin | | Descriptor: | F1845 fimbrial protein | | Authors: | Le Trong, I, Stenkamp, R.E, Korotkova, N, Moseley, S. | | Deposit date: | 2005-10-19 | | Release date: | 2005-11-01 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure and Mutational Analysis of the DaaE Adhesin of Escherichia coli.
J.Biol.Chem., 281, 2006
|
|
2D27
 
 | | Structure of the N-terminal domain of XpsE (crystal form I4122) | | Descriptor: | type II secretion ATPase XpsE | | Authors: | Chen, Y, Shiue, S.-J, Huang, C.-W, Chang, J.-L, Chien, Y.-L, Hu, N.-T, Chan, N.-L. | | Deposit date: | 2005-09-03 | | Release date: | 2005-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure and Function of the XpsE N-Terminal Domain, an Essential Component of the Xanthomonas campestris Type II Secretion System
J.Biol.Chem., 280, 2005
|
|
2BHV
 
 | | Structure of ComB10 of the Com Type IV secretion system of Helicobacter pylori | | Descriptor: | COMB10 | | Authors: | Terradot, L, Oomen, C, Bayliss, R, Leonard, G, Waksman, G. | | Deposit date: | 2005-01-18 | | Release date: | 2005-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of Two Core Subunits of the Bacterial Type Iv Secretion System, Virb8 from Brucella Suis and Comb10 from Helicobacter Pylori
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BS7
 
 | | Crystal structure of F17b-G in complex with chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, F17BG LECTIN | | Authors: | Buts, L, Wellens, A, Van Molle, I, Wyns, L, Loris, R, Lahmann, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2005-05-18 | | Release date: | 2006-05-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Impact of Natural Variation in Bacterial F17G Adhesins on Crystallization Behaviour.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2BSB
 
 | | E. coli F17e-G lectin domain complex with N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F17G ADHESIN SUBUNIT | | Authors: | Buts, L, Wellens, A, Van Molle, I, Wyns, L, Loris, R, Lahmann, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2005-05-20 | | Release date: | 2006-05-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Impact of Natural Variation in Bacterial F17G Adhesins on Crystallization Behaviour.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2BHM
 
 | | Crystal structure of VirB8 from Brucella suis | | Descriptor: | TYPE IV SECRETION SYSTEM PROTEIN VIRB8 | | Authors: | Bayliss, R, Baron, C, Waksman, G. | | Deposit date: | 2005-01-14 | | Release date: | 2005-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Two Core Subunits of the Bacterial Type Iv Secretion System, Virb8 from Brucella Suis and Comb10 from Helicobacter Pylori
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BS8
 
 | | Crystal structure of F17b-G in complex with N-acetyl-D-glucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADHESIN | | Authors: | Buts, L, Wellens, A, VanMolle, I, Wyns, L, Loris, R, Lahmann, M, Oscarson, S, DeGreve, H, Bouckaert, J. | | Deposit date: | 2005-05-18 | | Release date: | 2005-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Impact of Natural Variation in Bacterial F17G Adhesins on Crystallization Behaviour.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2BSC
 
 | | E. coli F17a-G lectin domain complex with N-acetylglucosamine, high- resolution structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F17A-G ADHESIN | | Authors: | Buts, L, Wellens, A, Van Molle, I, Wyns, L, Loris, R, Lahmann, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2005-05-20 | | Release date: | 2006-05-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Impact of Natural Variation in Bacterial F17G Adhesins on Crystallization Behaviour.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
8USM
 
 | | FmlH Lectin Domain UTI89 - AM4085 | | Descriptor: | 4'-fluoro-6-(trifluoromethyl)[1,1'-biphenyl]-2-yl 2-acetamido-2-deoxy-beta-D-galactopyranoside, Fimbrial protein (Fragment) | | Authors: | Tamadonfar, K.O, Pinkner, J.P, Maddirala, A.R, Janetka, J.W, Hultgren, S.J. | | Deposit date: | 2023-10-27 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery of Orally Bioavailable FmlH Lectin Antagonists as Treatment for Urinary Tract Infections.
J.Med.Chem., 67, 2024
|
|
8ENR
 
 | |
4KZ1
 
 | |
1ZK5
 
 | | Escherichia coli F17fG lectin domain complex with N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F17G adhesin subunit | | Authors: | Buts, L, Wellens, A, Van Molle, I, De Genst, E, Wyns, L, Loris, R, Lahmann, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2005-05-02 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Impact of natural variation in bacterial F17G adhesins on crystallization behaviour.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
8ENQ
 
 | | E. coli CsgA fibril (218-pixel box size) | | Descriptor: | Major curlin subunit | | Authors: | Bu, F, Liu, B. | | Deposit date: | 2022-09-30 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insight into Escherichia coli CsgA amyloid fibril assembly.
Mbio, 15, 2024
|
|
4JF8
 
 | |
4LFD
 
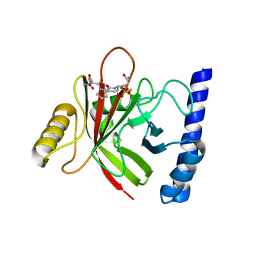 | | Staphylococcus aureus sortase B-substrate complex | | Descriptor: | (CBZ)NPQ(B27) PEPTIDE, SULFATE ION, Sortase B | | Authors: | Jacobitz, A.W, Sawaya, M.R, Yi, S.W, Amer, B.R, Huang, G.L, Nguyen, A.V, Jung, M.E, Clubb, R.T. | | Deposit date: | 2013-06-26 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural and Computational Studies of the Staphylococcus aureus Sortase B-Substrate Complex Reveal a Substrate-stabilized Oxyanion Hole.
J.Biol.Chem., 289, 2014
|
|
1ZPL
 
 | | E. coli F17a-G lectin domain complex with GlcNAc(beta1-O)Me | | Descriptor: | F17a-G, methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside | | Authors: | Buts, L, Wellens, A, Van Molle, I, Wyns, L, Loris, R, Lahmann, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2005-05-17 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Impact of natural variation in bacterial F17G adhesins on crystallization behaviour.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5F2F
 
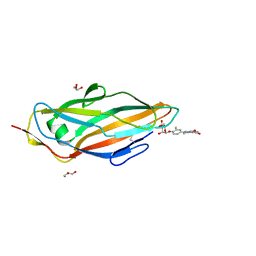 | | Crystal structure of para-biphenyl-2-methyl-3', 5' di-methyl amide mannoside bound to FimH lectin domain | | Descriptor: | 5-[4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3-methyl-phenyl]-~{N}1,~{N}3-dimethyl-benzene-1,3-dicarboxamide, GLYCEROL, Protein FimH | | Authors: | Kalas, V, Hultgren, S.J. | | Deposit date: | 2015-12-01 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Antivirulence Isoquinolone Mannosides: Optimization of the Biaryl Aglycone for FimH Lectin Binding Affinity and Efficacy in the Treatment of Chronic UTI.
Chemmedchem, 11, 2016
|
|
6K73
 
 | |
6KNY
 
 | |
6EF8
 
 | | Cryo-EM of the OmcS nanowires from Geobacter sulfurreducens | | Descriptor: | C-type cytochrome OmcS, HEME C | | Authors: | Wang, F, Gu, Y, Egelman, E.H, Malvankar, N.S. | | Deposit date: | 2018-08-16 | | Release date: | 2019-04-10 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of Microbial Nanowires Reveals Stacked Hemes that Transport Electrons over Micrometers.
Cell, 177, 2019
|
|
4NHF
 
 | |
3SNK
 
 | |
3T1B
 
 | | Crystal structure of the full-length AphB N100E variant | | Descriptor: | Transcriptional regulator, LysR family | | Authors: | Taylor, J.L, De Silva, R.S, Kovacikova, G, Lin, W, Taylor, R.K, Skorupski, K, Kull, F.J. | | Deposit date: | 2011-07-21 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of AphB, a virulence gene activator from Vibrio cholerae, reveals residues that influence its response to oxygen and pH.
Mol.Microbiol., 83, 2012
|
|
