5GRV
 
 | | Crystal structure of homo-specific diabody | | Descriptor: | homo-specific diabody heavy chain, homo-specific diabody light chain | | Authors: | Kim, J.H, Song, D.H, Youn, S.J, Kim, J.W, Cho, G, Lee, H, Lee, J.O. | | Deposit date: | 2016-08-12 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of mono- and bi-specific diabodies and reduction of their structural flexibility by introduction of disulfide bridges at the Fv interface.
Sci Rep, 6, 2016
|
|
4AL0
 
 | | Crystal structure of Human PS-1 | | Descriptor: | GLUTATHIONE, PALMITIC ACID, PROSTAGLANDIN E SYNTHASE, ... | | Authors: | Sjogren, T, Nord, J, Ek, M, Johansson, P, Liu, G, Geschwindner, S. | | Deposit date: | 2012-02-29 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal Structure of Microsomal Prostaglandin E2 Synthase Provides Insight Into Diversity in the Mapeg Superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5GS3
 
 | | Crystal structure of diabody | | Descriptor: | diabody protein | | Authors: | Kim, J.H, Song, D.H, Youn, S.J, Kim, J.W, Cho, G, Lee, H, Lee, J.O. | | Deposit date: | 2016-08-13 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of mono- and bi-specific diabodies and reduction of their structural flexibility by introduction of disulfide bridges at the Fv interface.
Sci Rep, 6, 2016
|
|
5GRX
 
 | | Crystal structure of disulfide-bonded diabody | | Descriptor: | diabody protein | | Authors: | Kim, J.H, Song, D.H, Youn, S.J, Kim, J.W, Cho, G, Lee, H, Lee, J.O. | | Deposit date: | 2016-08-12 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal structure of mono- and bi-specific diabodies and reduction of their structural flexibility by introduction of disulfide bridges at the Fv interface.
Sci Rep, 6, 2016
|
|
5NCM
 
 | | Crystal structure Cbk1(NTR)-Mob2 complex | | Descriptor: | CBK1 kinase activator protein MOB2, Serine/threonine-protein kinase CBK1, ZINC ION | | Authors: | Gogl, G, Remenyi, A, Parker, B, Weiss, E. | | Deposit date: | 2017-03-06 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ndr/Lats Kinases Bind Specific Mob-Family Coactivators through a Conserved and Modular Interface.
Biochemistry, 2020
|
|
5KF3
 
 | |
5KEH
 
 | |
5KKD
 
 | |
8G0P
 
 | |
8G0Q
 
 | |
1O7T
 
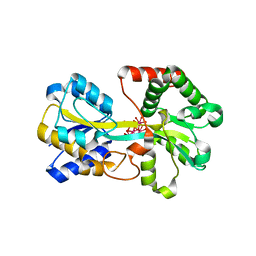 | | Metal nanoclusters bound to the Ferric Binding Protein from Neisseria gonorrhoeae. | | Descriptor: | HF OXO CLUSTER HF5, HF-OXO-PHOSPHATE CLUSTER PHF, IRON BINDING PROTEIN, ... | | Authors: | Alexeev, D, Zu, H, Guo, M, Zhong, W, Hunter, D.J.B, Yang, W, Campopiano, D.J, Sadler, P.J. | | Deposit date: | 2002-11-12 | | Release date: | 2003-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A novel protein-mineral interface.
Nat. Struct. Biol., 10, 2003
|
|
5NCN
 
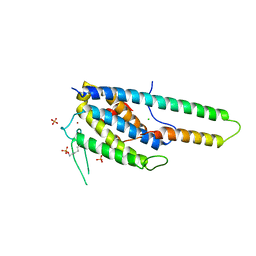 | | Crystal structure Dbf2(NTR)-Mob1 complex | | Descriptor: | CHLORIDE ION, Cell cycle protein kinase DBF2, DBF2 kinase activator protein MOB1, ... | | Authors: | Gogl, G, Remenyi, A, Parker, B, Weiss, E. | | Deposit date: | 2017-03-06 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Ndr/Lats Kinases Bind Specific Mob-Family Coactivators through a Conserved and Modular Interface.
Biochemistry, 2020
|
|
5GRZ
 
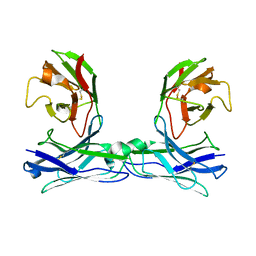 | | Crystal structure of disulfide-bonded diabody | | Descriptor: | diabody | | Authors: | Kim, J.H, Song, D.H, Youn, S.J, Kim, J.W, Cho, G, Lee, H, Lee, J.O. | | Deposit date: | 2016-08-13 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of mono- and bi-specific diabodies and reduction of their structural flexibility by introduction of disulfide bridges at the Fv interface.
Sci Rep, 6, 2016
|
|
4EHY
 
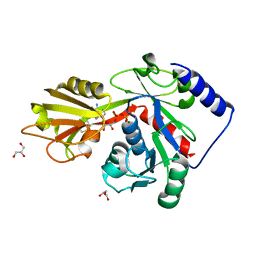 | | Crystal structure of LpxK from Aquifex aeolicus in complex with ADP/Mg2+ at 2.2 angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Emptage, R.P, Daughtry, K.D, Pemble IV, C.W, Raetz, C.R.H. | | Deposit date: | 2012-04-04 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Crystal structure of LpxK, the 4'-kinase of lipid A biosynthesis and atypical P-loop kinase functioning at the membrane interface.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4EHW
 
 | | Crystal structure of LpxK from Aquifex aeolicus at 2.3 angstrom resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Tetraacyldisaccharide 4'-kinase | | Authors: | Emptage, R.P, Daughtry, K.D, Pemble IV, C.W, Raetz, C.R.H. | | Deposit date: | 2012-04-04 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of LpxK, the 4'-kinase of lipid A biosynthesis and atypical P-loop kinase functioning at the membrane interface.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4EHX
 
 | | Crystal structure of LpxK from Aquifex aeolicus at 1.9 angstrom resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Emptage, R.P, Daughtry, K.D, Pemble IV, C.W, Raetz, C.R.H. | | Deposit date: | 2012-04-04 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of LpxK, the 4'-kinase of lipid A biosynthesis and atypical P-loop kinase functioning at the membrane interface.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5XOR
 
 | |
7MQY
 
 | | Zebrafish CNTN4 APLP2 complex | | Descriptor: | Fusion protein of CNTN4 and APLP2 | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
7MRQ
 
 | |
6MI3
 
 | | Structure of NEMO(51-112) with N- and C-terminal coiled-coil adaptors. | | Descriptor: | NF-kB ESSENTIAL MODULATOR,NF-kappa-B essential modulator,NF-kB ESSENTIAL MODULATOR | | Authors: | Pellegrini, M, Barczewski, A.H, Mierke, D.F, Ragusa, M.J. | | Deposit date: | 2018-09-19 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | The IKK-binding domain of NEMO is an irregular coiled coil with a dynamic binding interface.
Sci Rep, 9, 2019
|
|
6MI4
 
 | |
7MRO
 
 | | Zebrafish CNTN4 FN1-FN3 domains | | Descriptor: | Contactin-4 | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
7MRS
 
 | | Zebrafish CNTN4 APPb complex | | Descriptor: | Amyloid-beta A4 protein, Contactin-4 | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-09 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
7MRM
 
 | | Chicken CNTN3 APP complex | | Descriptor: | Fusion protein of Chicken CNTN3 FN1-FN2 domains and Amyloid-beta A4 protein,Amyloid-beta A4 protein, SULFATE ION | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
7MRK
 
 | | Chicken CNTN4 APP complex | | Descriptor: | Amyloid-beta A4 protein, Contactin-4, DI(HYDROXYETHYL)ETHER | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
