3K05
 
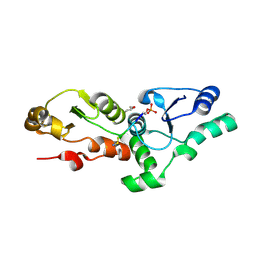 | |
3CV0
 
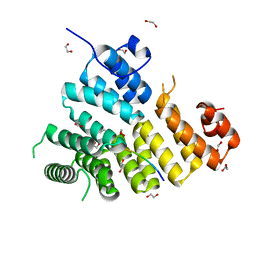 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to T. brucei Phosphoglucoisomerase (PGI) PTS1 peptide | | Descriptor: | 1,2-ETHANEDIOL, Peroxisome targeting signal 1 receptor PEX5, T. brucei PGI PTS1 peptide Ac-FNELSHL | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-17 | | Release date: | 2008-06-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
3K0K
 
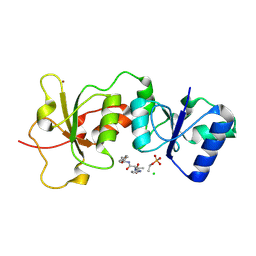 | | Crystal Structure of BRCA1 BRCT in complex with a minimal recognition tetrapeptide with a free carboxy C-terminus. | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
3DDK
 
 | | Coxsackievirus B3 3Dpol RNA Dependent RNA Polymerase | | Descriptor: | RNA polymerase B3 3Dpol, SODIUM ION, SULFATE ION | | Authors: | Campagnola, G, Weygandt, M.H, Scoggin, K.E, Peersen, O.B. | | Deposit date: | 2008-06-05 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Coxsackievirus B3 3Dpol Highlights Functional Importance of Residue 5 in Picornaviral Polymerases
J.Virol., 82, 2008
|
|
2IP4
 
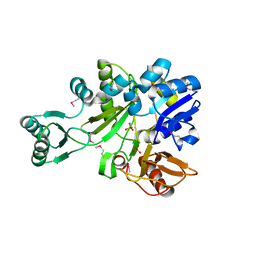 | | Crystal Structure of Glycinamide Ribonucleotide Synthetase from Thermus thermophilus HB8 | | Descriptor: | Phosphoribosylamine--glycine ligase, SULFATE ION | | Authors: | Sampei, G, Baba, S, Kanagawa, M, Yanai, H, Ishii, T, Kawai, H, Fukai, Y, Ebihara, A, Nakagawa, N, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-11 | | Release date: | 2007-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
1TVZ
 
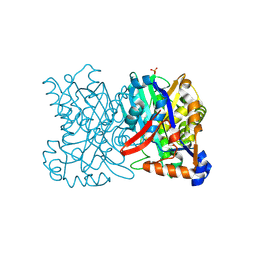 | | Crystal structure of 3-hydroxy-3-methylglutaryl-coenzyme A synthase from Staphylococcus aureus | | Descriptor: | 3-hydroxy-3-methylglutaryl-CoA synthase, SULFATE ION | | Authors: | Campobasso, N, Patel, M, Wilding, I.E, Kallender, H, Rosenberg, M, Gwynn, M. | | Deposit date: | 2004-06-30 | | Release date: | 2004-08-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase: crystal structure and mechanism
J.Biol.Chem., 279, 2004
|
|
1KI4
 
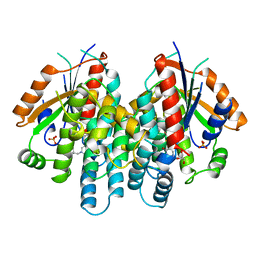 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH 5-BROMOTHIENYLDEOXYURIDINE | | Descriptor: | 5-BROMOTHIENYLDEOXYURIDINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Visse, R, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-18 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
1KU7
 
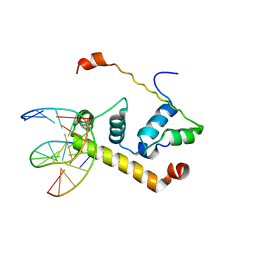 | | Crystal Structure of Thermus aquatics RNA Polymerase SigmaA Subunit Region 4 Bound to-35 Element DNA | | Descriptor: | 5'-D(*CP*CP*TP*TP*GP*AP*CP*AP*AP*AP*G)-3', 5'-D(*CP*CP*TP*TP*TP*GP*TP*CP*AP*AP*G)-3', sigma factor sigA | | Authors: | Campbell, E.A, Muzzin, O, Chlenov, M, Sun, J.L, Olson, C.A, Weinman, O, Trester-Zedlitz, M.L, Darst, S.A. | | Deposit date: | 2002-01-21 | | Release date: | 2002-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the bacterial RNA polymerase promoter specificity sigma subunit.
Mol.Cell, 9, 2002
|
|
1KU3
 
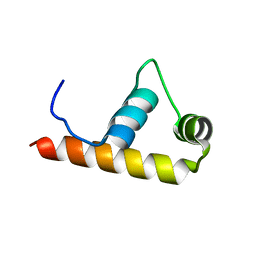 | | Crystal Structure of Thermus aquaticus RNA Polymerase Sigma Subunit Fragment, Region 4 | | Descriptor: | sigma factor sigA | | Authors: | Campbell, E.A, Muzzin, O, Chlenov, M, Sun, J.L, Olson, C.A, Weinman, O, Trester-Zedlitz, M.L, Darst, S.A. | | Deposit date: | 2002-01-21 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the bacterial RNA polymerase promoter specificity sigma subunit.
Mol.Cell, 9, 2002
|
|
1L0O
 
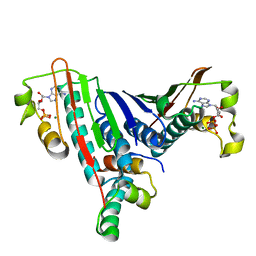 | | Crystal Structure of the Bacillus stearothermophilus Anti-Sigma Factor SpoIIAB with the Sporulation Sigma Factor SigmaF | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Anti-sigma F factor, MAGNESIUM ION, ... | | Authors: | Campbell, E.A, Masuda, S, Sun, J.L, Muzzin, O, Olson, C.A, Wang, S, Darst, S.A. | | Deposit date: | 2002-02-12 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the Bacillus stearothermophilus anti-sigma factor SpoIIAB with the sporulation sigma factor sigmaF.
Cell(Cambridge,Mass.), 108, 2002
|
|
8B02
 
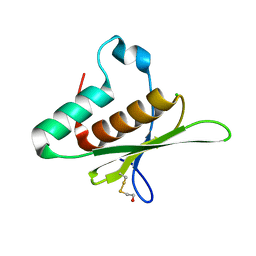 | | Crystal structure of the dsRBD domain of tRNA-dihydrouridine(20) synthase from Amphimedon queenslandica | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, DRBM domain-containing protein, ... | | Authors: | Pecqueur, L, Faivre, B, Hamdane, D. | | Deposit date: | 2022-09-07 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.676 Å) | | Cite: | Evolutionary Diversity of Dus2 Enzymes Reveals Novel Structural and Functional Features among Members of the RNA Dihydrouridine Synthases Family.
Biomolecules, 12, 2022
|
|
8AR9
 
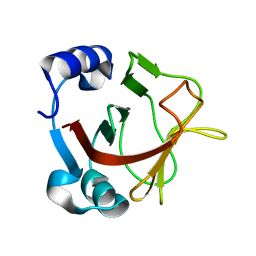 | |
8AR6
 
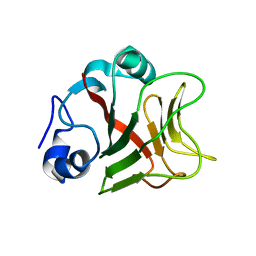 | |
6P3E
 
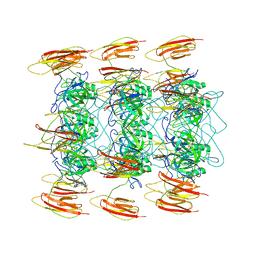 | | Mobile loops and electrostatic interactions maintain the flexible lambda tail tube | | Descriptor: | Tail tube protein | | Authors: | Campbell, P, Duda, R.L, Nassur, J, Hendrix, R.W, Conway, J.F, Huet, A. | | Deposit date: | 2019-05-23 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Mobile Loops and Electrostatic Interactions Maintain the Flexible Tail Tube of Bacteriophage Lambda.
J.Mol.Biol., 432, 2020
|
|
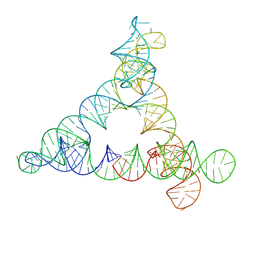
8BTZ
 
 | |
8BU8
 
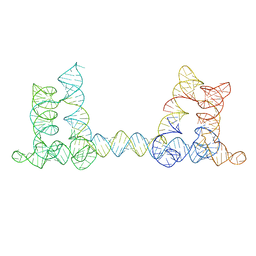 | |
9NR7
 
 | |
4UP1
 
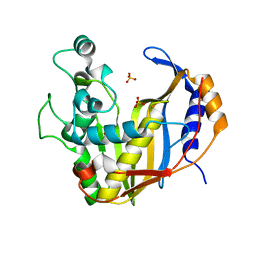 | |
4UOR
 
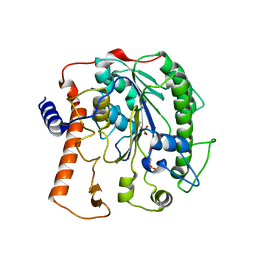 | | Structure of lipoteichoic acid synthase LtaS from Listeria monocytogenes in complex with glycerol phosphate | | Descriptor: | (2R)-2,3-dihydroxypropyl phosphate, LIPOTEICHOIC ACID SYNTHASE, MAGNESIUM ION | | Authors: | Campeotto, I, Freemont, P, Grundling, A. | | Deposit date: | 2014-06-09 | | Release date: | 2014-08-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural and mechanistic insight into the Listeria monocytogenes two-enzyme lipoteichoic acid synthesis system.
J. Biol. Chem., 289, 2014
|
|
4UOP
 
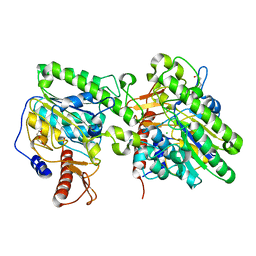 | | Crystal structure of the lipoteichoic acid synthase LtaP from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, LIPOTEICHOIC ACID PRIMASE, MAGNESIUM ION, ... | | Authors: | Campeotto, I, Freemont, P, Grundling, A. | | Deposit date: | 2014-06-06 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and mechanistic insight into the Listeria monocytogenes two-enzyme lipoteichoic acid synthesis system.
J. Biol. Chem., 289, 2014
|
|
4UOO
 
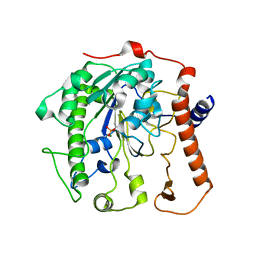 | |
5I22
 
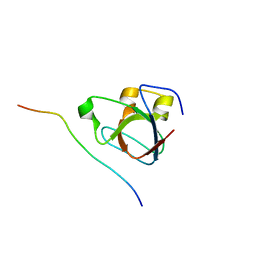 | | Amphiphysin SH3 in complex with Chikungunya virus nsP3 peptide | | Descriptor: | CHIKV nsP3 peptide, Myc box-dependent-interacting protein 1 | | Authors: | Tossavainen, H, Aitio, O, Hellman, M, Saksela, K, Permi, P. | | Deposit date: | 2016-02-04 | | Release date: | 2016-06-15 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the High Affinity Interaction between the Alphavirus Nonstructural Protein-3 (nsP3) and the SH3 Domain of Amphiphysin-2.
J.Biol.Chem., 291, 2016
|
|
5IDE
 
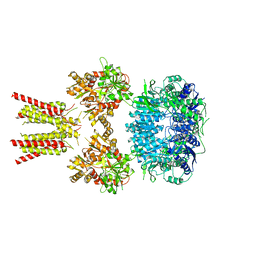 | | Cryo-EM structure of GluA2/3 AMPA receptor heterotetramer (model I) | | Descriptor: | Glutamate receptor 2, Glutamate receptor 3 | | Authors: | Herguedas, B, Garcia-Nafria, J, Fernandez-Leiro, R, Greger, I.H. | | Deposit date: | 2016-02-24 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.25 Å) | | Cite: | Structure and organization of heteromeric AMPA-type glutamate receptors.
Science, 352, 2016
|
|
5IDF
 
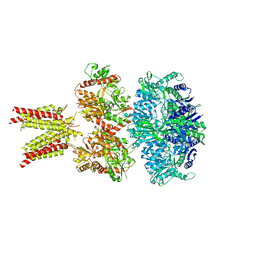 | | Cryo-EM structure of GluA2/3 AMPA receptor heterotetramer (model II) | | Descriptor: | Glutamate receptor 2, Glutamate receptor 3 | | Authors: | Herguedas, B, Garcia-Nafria, J, Fernandez-Leiro, R, Greger, I.H. | | Deposit date: | 2016-02-24 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (10.31 Å) | | Cite: | Structure and organization of heteromeric AMPA-type glutamate receptors.
Science, 352, 2016
|
|
6Z4K
 
 | |
