6A82
 
 | |
6ADZ
 
 | | LdCoroCC Double mutant- I486A-L493A | | Descriptor: | Coronin-like protein, SULFATE ION | | Authors: | Karade, S.S, Ansari, A, Pratap, J.V. | | Deposit date: | 2018-08-02 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.431 Å) | | Cite: | Molecular and structural analysis of a mechanical transition of helices in the L. donovani coronin coiled-coil domain.
Int.J.Biol.Macromol., 143, 2020
|
|
6AJD
 
 | |
6EVG
 
 | |
6B0D
 
 | | An E. coli DPS protein from ferritin superfamily | | Descriptor: | DNA protection during starvation protein, FORMIC ACID, SODIUM ION | | Authors: | Rui, W, Ruslan, S, Ronan, K, Adam, J.S. | | Deposit date: | 2017-09-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | SIMBAD: a sequence-independent molecular-replacement pipeline.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6GOA
 
 | |
5WT4
 
 | | L-Cysteine-PLP intermediate of NifS from Helicobacter pylori | | Descriptor: | Cysteine desulfurase IscS, ISOPROPYL ALCOHOL, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-CYSTEINE | | Authors: | Fujishiro, T, Nakamura, R, Takahashi, T. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Snapshots of PLP-substrate and PLP-product external aldimines as intermediates in two types of cysteine desulfurase enzymes.
Febs J., 2019
|
|
1UPN
 
 | | COMPLEX OF ECHOVIRUS TYPE 12 WITH DOMAINS 3 AND 4 OF ITS RECEPTOR DECAY ACCELERATING FACTOR (CD55) BY CRYO ELECTRON MICROSCOPY AT 16 A | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR, ECHOVIRUS 11 COAT PROTEIN VP1, ECHOVIRUS 11 COAT PROTEIN VP2, ... | | Authors: | Bhella, D, Goodfellow, I.G, Roversi, P, Pettigrew, D, Chaudry, Y, Evans, D.J, Lea, S.M. | | Deposit date: | 2003-10-08 | | Release date: | 2004-01-07 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | The Structure of Echovirus Type 12 Bound to a Two-Domain Fragment of its Cellular Attachment Protein Decay-Accelerating Factor (Cd 55)
J.Biol.Chem., 279, 2004
|
|
1VIU
 
 | |
6B6M
 
 | | Cyanase from Serratia proteamaculans | | Descriptor: | Cyanate hydratase | | Authors: | Xu, Y. | | Deposit date: | 2017-10-02 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | SIMBAD: a sequence-independent molecular-replacement pipeline.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
1VHD
 
 | |
1VHW
 
 | |
1VHJ
 
 | |
6BY0
 
 | |
5HKE
 
 | | bile salt hydrolase from Lactobacillus salivarius | | Descriptor: | Bile salt hydrolase, PHOSPHATE ION | | Authors: | Hu, X.-J. | | Deposit date: | 2016-01-14 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of bile salt hydrolase from Lactobacillus salivarius.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5YIA
 
 | | Crystal Structure of KNI-10343 bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-[[(2R)-2-[2-(4-hydroxyphenyl)ethanoylamino]-3-methylsulfanyl-propanoyl]amino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, GLYCEROL, ... | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2017-10-03 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Febs J., 285, 2018
|
|
1TKQ
 
 | | SOLUTION STRUCTURE OF A LINKED UNSYMMETRIC GRAMICIDIN IN A MEMBRANE-ISOELECTRICAL SOLVENTS MIXTURE IN THE PRESENCE OF CsCl | | Descriptor: | GRAMICIDIN A, MINI-GRAMICIDIN A, SUCCINIC ACID | | Authors: | Xie, X, Al-Momani, L, Bockelmann, D, Griesinger, C, Koert, U. | | Deposit date: | 2004-06-09 | | Release date: | 2004-07-13 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | An Asymmetric Ion Channel Derived from Gramicidin A. Synthesis, Function and NMR Structure.
FEBS J., 272, 2005
|
|
1VIZ
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | PcrB protein homolog, SODIUM ION | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
5HMJ
 
 | |
5YIE
 
 | | Crystal Structure of KNI-10742 bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-[2-[4-[2-azanylethyl(ethyl)amino]-2,6-dimethyl-phenoxy]ethanoylamino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Plasmepsin II, ... | | Authors: | Mishra, V, Rathore, I, Bhaumik, P. | | Deposit date: | 2017-10-04 | | Release date: | 2018-07-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Febs J., 285, 2018
|
|
5AN1
 
 | |
6G6R
 
 | | Human Methionine Adenosyltransferase II with SAMe and PPNP | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Panmanee, J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2018-04-02 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory beta subunit and quinolone-based compounds.
Febs J., 286, 2019
|
|
5YIC
 
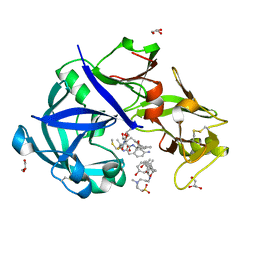 | | Crystal Structure of KNI-10333 bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-[[(2R)-2-[2-(4-aminophenyl)ethanoylamino]-3-methylsulfanyl-propanoyl]amino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, GLYCEROL, ... | | Authors: | Mishra, V, Rathore, I, Bhaumik, P. | | Deposit date: | 2017-10-03 | | Release date: | 2018-07-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Febs J., 285, 2018
|
|
1W6B
 
 | | Solution NMR Structure of a Long Neurotoxin from the Venom of the Asian Cobra, 20 Structures | | Descriptor: | LONG NEUROTOXIN 1 | | Authors: | Talebzadeh-Farooji, M, Amininasab, M, Elmi, M.M, Naderi-Manesh, H, Sarbolouki, M.N. | | Deposit date: | 2004-08-17 | | Release date: | 2004-12-22 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of long neurotoxin NTX-1 from the venom of Naja naja oxiana by 2D-NMR spectroscopy.
Eur. J. Biochem., 271, 2004
|
|
6MVH
 
 | |
