2W0U
 
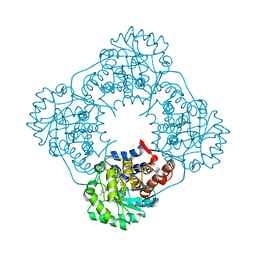 | | CRYSTAL STRUCTURE OF HUMAN GLYCOLATE OXIDASE IN COMPLEX WITH THE INHIBITOR 5-[(4-CHLOROPHENYL)SULFANYL]- 1,2,3-THIADIAZOLE-4-CARBOXYLATE. | | Descriptor: | 5-[(4-chlorophenyl)sulfanyl]-1,2,3-thiadiazole-4-carboxylate, FLAVIN MONONUCLEOTIDE, HYDROXYACID OXIDASE 1 | | Authors: | Bourhis, J.M, Lindqvist, Y. | | Deposit date: | 2008-10-10 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure of Human Glycolate Oxidase in Complex with the Inhibitor 4-Carboxy-5-[(4-Chlorophenyl)Sulfanyl]-1,2,3-Thiadiazole.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
5UGA
 
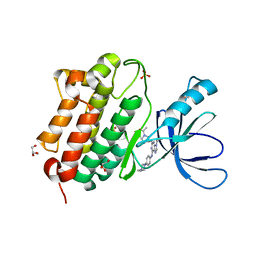 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with 4-(4-{[2-{[(3S)-1-acetylpyrrolidin-3-yl]amino}-9-(propan-2-yl)-9H-purin-6-yl]amino}phenyl)-1-methylpiperazin-1-ium | | Descriptor: | 4-(4-{[2-{[(3S)-1-acetylpyrrolidin-3-yl]amino}-9-(propan-2-yl)-9H-purin-6-yl]amino}phenyl)-1-methylpiperazin-1-ium, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S, Ferre, R.A. | | Deposit date: | 2017-01-07 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
5UGB
 
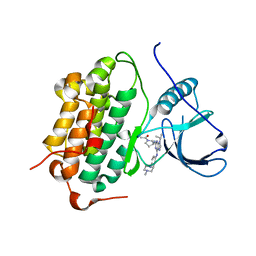 | |
4CM8
 
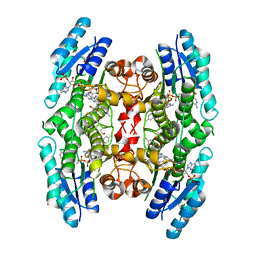 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | (E)-2,4-diamino-6-(4-methylstyryl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4CM5
 
 | |
5DUL
 
 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis in complex with NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Osipiuk, J, Mulligan, R, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis in complex with NADPH .
to be published
|
|
2Y1F
 
 | | X-ray structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with a 3,4- dichlorophenyl-substituted fosmidomycin analogue, manganese and NADPH. | | Descriptor: | (1S)-1-(3,4-DICHLOROPHENYL)-3-[FORMYL(HYDROXY)AMINO]PROPYL}PHOSPHONIC ACID, 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, MANGANESE (II) ION, ... | | Authors: | Henriksson, L.M, Larsson, A.M.S, Bergfors, T, Bjorkelid, C, Unge, T, Mowbray, S.L, Jones, T.A. | | Deposit date: | 2010-12-08 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Design, Synthesis and X-Ray Crystallographic Studies of Alpha-Aryl Substituted Fosmidomycin Analogues as Inhibitors of Mycobacterium Tuberculosis 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase
J.Med.Chem, 54, 2011
|
|
2X2J
 
 | | Crystal structure of the Gracilariopsis lemaneiformis alpha- 1,4-glucan lyase with deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, CHLORIDE ION, ... | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
5U6V
 
 | | X-ray crystal structure of 1,2,3-triazolobenzodiazepine in complex with BRD2(D2) | | Descriptor: | 1,2-ETHANEDIOL, 5-[7-(4-chlorophenyl)-1-methyl-6,7-dihydro-5H-[1,2,3]triazolo[1,5-d][1,4]benzodiazepin-9-yl]pyridin-2-amine, Bromodomain-containing protein 2 | | Authors: | Hatfaludi, T, Sharp, P.P, Garnier, J.-M, Burns, C.J, Czabotar, P.E. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.775 Å) | | Cite: | Design, Synthesis, and Biological Activity of 1,2,3-Triazolobenzodiazepine BET Bromodomain Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
2Y1D
 
 | | X-ray structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with a 3,4- dichlorophenyl-substituted fosmidomycin analogue and manganese. | | Descriptor: | (1S)-1-(3,4-DICHLOROPHENYL)-3-[FORMYL(HYDROXY)AMINO]PROPYL}PHOSPHONIC ACID, 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, MANGANESE (II) ION | | Authors: | Henriksson, L.M, Larsson, A.M.S, Bergfors, T, Bjorkelid, C, Unge, T, Mowbray, S.L, Jones, T.A. | | Deposit date: | 2010-12-08 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design, Synthesis and X-Ray Crystallographic Studies of Alpha-Aryl Substituted Fosmidomycin Analogues as Inhibitors of Mycobacterium Tuberculosis 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase
J.Med.Chem, 54, 2011
|
|
5Z5R
 
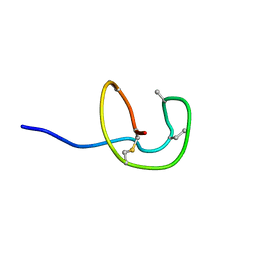 | | Nukacin ISK-1 in inactive state | | Descriptor: | Lantibiotic nukacin | | Authors: | Kohda, D, Fujinami, D. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | The lantibiotic nukacin ISK-1 exists in an equilibrium between active and inactive lipid-II binding states.
Commun Biol, 1, 2018
|
|
5DU4
 
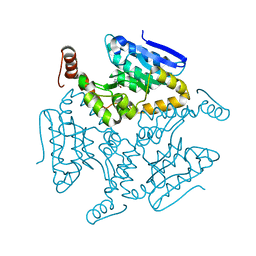 | | Crystal structure of M. tuberculosis EchA6 bound to ligand GSK366A | | Descriptor: | (5R,7S)-5-(4-ethylphenyl)-N-(4-methoxybenzyl)-7-(trifluoromethyl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
4CLO
 
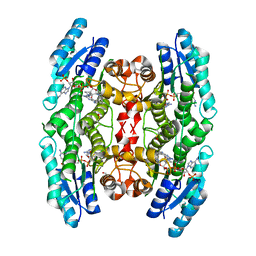 | |
2YCE
 
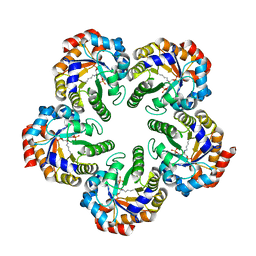 | | Structure of an Archaeal fructose-1,6-bisphosphate aldolase with the catalytic Lys covalently bound to the carbinolamine intermediate of the substrate. | | Descriptor: | D-MANNITOL-1,6-DIPHOSPHATE, FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS 1 | | Authors: | Lorentzen, E, Siebers, B, Hensel, R, Pohl, E. | | Deposit date: | 2011-03-14 | | Release date: | 2011-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanism of the Schiff Base Forming Fructose-1,6-Bisphosphate Aldolase: Structural Analysis of Reaction Intermediates.
Biochemistry, 44, 2005
|
|
2WMV
 
 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1-[(2S)-4-(7H-PURIN-6-YL)MORPHOLIN-2-YL]METHANAMINE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, Klair, S, Burns, S, Boxall, K, Cherry, M, Fisher, M, Westwood, I.M, Walton, M.I, McHardy, T, Cheung, K.-M.J, Van Montfort, R, Williams, D, Aherne, G.W, Garrett, M.D, Reader, J, Collins, I. | | Deposit date: | 2009-07-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Identification of Inhibitors of Checkpoint Kinase 1 Through Template Screening.
J.Med.Chem., 52, 2009
|
|
5TYS
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-142 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2016-11-21 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | A novel central nervous system-penetrating protease inhibitor overcomes human immunodeficiency virus 1 resistance with unprecedented aM to pM potency.
Elife, 6, 2017
|
|
3IV7
 
 | |
5T8K
 
 | | 1.95 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Adenine and NAD | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-07 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Adenine and NAD.
To Be Published
|
|
3K31
 
 | |
2VNT
 
 | | Urokinase-Type Plasminogen Activator Inhibitor Complex with a 1-(7- SULPHOAMIDOISOQUINOLINYL)GUANIDINE | | Descriptor: | 1-({4-CHLORO-1-[(DIAMINOMETHYLIDENE)AMINO]ISOQUINOLIN-7-YL}SULFONYL)-D-PROLINE, SULFATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Fish, P.V, Barber, C.G, Brown, D.G, Butt, R, Henry, B.T, Horne, V, Huggins, J.P, McCleverty, D, Phillips, C, Webster, R, Dickinson, R.P, Collis, M.G, King, E, O'Gara, M, McIntosh, F. | | Deposit date: | 2008-02-07 | | Release date: | 2008-02-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selective Urokinase-Type Plasminogen Activator (Upa) Inhibitors 4. 1-(7-Sulphonamidoisoquinolinyl) Guanidines
J.Med.Chem., 50, 2007
|
|
5TYR
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-121 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2016-11-21 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel central nervous system-penetrating protease inhibitor overcomes human immunodeficiency virus 1 resistance with unprecedented aM to pM potency.
Elife, 6, 2017
|
|
5V36
 
 | | 1.88 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus mutans UA159 in Complex with FAD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-03-06 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus mutans UA159 in Complex with FAD.
To Be Published
|
|
7JHO
 
 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-21 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
5DU8
 
 | | Crystal structure of M. tuberculosis EchA6 bound to GSK572A | | Descriptor: | (5R,7S)-5-(4-ethylphenyl)-N-[(5-fluoropyridin-2-yl)methyl]-7-(trifluoromethyl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
5TIM
 
 | | REFINED 1.83 ANGSTROMS STRUCTURE OF TRYPANOSOMAL TRIOSEPHOSPHATE ISOMERASE, CRYSTALLIZED IN THE PRESENCE OF 2.4 M-AMMONIUM SULPHATE. A COMPARISON WITH THE STRUCTURE OF THE TRYPANOSOMAL TRIOSEPHOSPHATE ISOMERASE-GLYCEROL-3-PHOSPHATE COMPLEX | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Wierenga, R.K, Hol, W.G.J. | | Deposit date: | 1991-04-23 | | Release date: | 1992-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Refined 1.83 A structure of trypanosomal triosephosphate isomerase crystallized in the presence of 2.4 M-ammonium sulphate. A comparison with the structure of the trypanosomal triosephosphate isomerase-glycerol-3-phosphate complex.
J.Mol.Biol., 220, 1991
|
|
