8UET
 
 | | In-situ complex I, Deactive class02 | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Zheng, W, Zhu, J, Zhang, K. | | Deposit date: | 2023-10-02 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 2024
|
|
8UES
 
 | | In-situ complex I, Deactive class01 | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Zheng, W, Zhu, J, Zhang, K. | | Deposit date: | 2023-10-02 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 2024
|
|
8UER
 
 | | In-situ complex I with Q10 (State-gamma) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Zheng, W, Zhu, J, Zhang, K. | | Deposit date: | 2023-10-02 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 2024
|
|
8UEQ
 
 | | In-situ complex I with Q10 (State-beta) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Zheng, W, Zhu, J, Zhang, K. | | Deposit date: | 2023-10-02 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 2024
|
|
8UEP
 
 | | In-situ complex I, Active-Q10 (State-alpha) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Zheng, W, Zhu, J, Zhang, K. | | Deposit date: | 2023-10-02 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 2024
|
|
8UEO
 
 | | In-situ complex I (Active-Apo) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Zheng, W, Zhu, J, Zhang, K. | | Deposit date: | 2023-10-02 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 2024
|
|
8UDV
 
 | | The X-RAY co-crystal structure of human FGFR3 V555M and Compound 17 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(6-chloro-1-cyclopropyl-1H-benzimidazol-5-yl)ethynyl]-1-[(3S,5S)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, Fibroblast growth factor receptor 3, ... | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
8UDU
 
 | | The X-RAY co-crystal structure of human FGFR3 and Compound 17 | | Descriptor: | 3-[(6-chloro-1-cyclopropyl-1H-benzimidazol-5-yl)ethynyl]-1-[(3S,5S)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, CHLORIDE ION, Fibroblast growth factor receptor 3 | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
8UDT
 
 | | The X-RAY co-crystal structure of human FGFR3 and KIN-3248 | | Descriptor: | 3-[(1-cyclopropyl-4,6-difluoro-1H-benzimidazol-5-yl)ethynyl]-1-[(3R,5R)-5-(methoxymethyl)-1-propanoylpyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, D-MALATE, Fibroblast growth factor receptor 3 | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.829 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
8UD1
 
 | | High resolution in-situ structure of complex I in respiratory supercomplex (composite) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Zheng, W, Zhu, J, Zhang, K. | | Deposit date: | 2023-09-27 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 2024
|
|
8UCI
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus K238G + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP dependent DNA ligase, GLYCEROL, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCH
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus K92A + ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP dependent DNA ligase, GLYCEROL, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCG
 
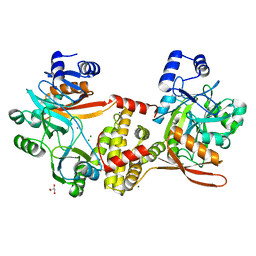 | | Thermophilic RNA Ligase from Palaeococcus pacificus K92A | | Descriptor: | ATP dependent DNA ligase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCF
 
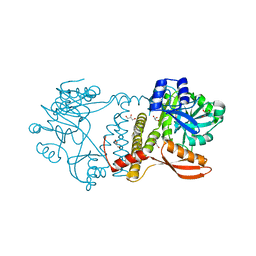 | | Thermophilic RNA Ligase from Palaeococcus pacificus K238G | | Descriptor: | ATP dependent DNA ligase, GLYCEROL, MAGNESIUM ION | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCE
 
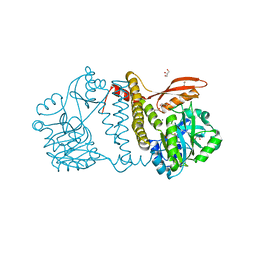 | | Thermophilic RNA Ligase from Palaeococcus pacificus + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP dependent DNA ligase, GLYCEROL, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCC
 
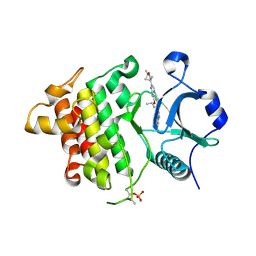 | | IRAK4 in complex with compound 20 | | Descriptor: | (4S)-2-[(1R,4s)-1-methyl-2-oxabicyclo[2.1.1]hexan-4-yl]-N-[(8S)-6-methylpyrazolo[1,5-a]pyrimidin-3-yl]-7-[(propan-2-yl)oxy]imidazo[1,2-a]pyrimidine-6-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Metrick, C.M, Chodaparambil, J.V. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Discovery of 7-Isopropoxy-2-(1-methyl-2-oxabicyclo[2.1.1]hexan-4-yl)- N -(6-methylpyrazolo[1,5- a ]pyrimidin-3-yl)imidazo[1,2- a ]pyrimidine-6-carboxamide (BIO-7488), a Potent, Selective, and CNS-Penetrant IRAK4 Inhibitor for the Treatment of Ischemic Stroke.
J.Med.Chem., 67, 2024
|
|
8UC6
 
 | | Calpain-7:IST1 Complex | | Descriptor: | Calpain-7, IST1 homolog | | Authors: | Paine, E, Whitby, F.G, Hill, C.P, Sunquist, W.I. | | Deposit date: | 2023-09-25 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | The Calpain-7 protease functions together with the ESCRT-III protein IST1 within the midbody to regulate the timing and completion of abscission.
Elife, 12, 2023
|
|
8UC1
 
 | |
8UAT
 
 | | Thermus scotoductus SA-01 Ene-reductase Compound 3b Complex | | Descriptor: | 1-[2-(4-hydroxyphenyl)ethyl]-1,4-dihydropyridine-3-carboxamide, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wilson, L.A, Guddat, L.W, Schenk, G, Scott, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural Characterization of Enzymatic Interactions with Functional Nicotinamide Cofactor Biomimetics
Catalysts, 14, 2024
|
|
8UAS
 
 | | Rhodococcus ruber Alcohol Dehydrogenase NADH Biomimetic Complex - Compound 1a | | Descriptor: | 1-[3-[~{tert}-butyl(dimethyl)silyl]oxypropyl]pyridine-3-carboxamide, CITRIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Wilson, L.A, Guddat, L.W, Schenk, G, Scott, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Characterization of Enzymatic Interactions with Functional Nicotinamide Cofactor Biomimetics
Catalysts, 14, 2024
|
|
8UAR
 
 | | Rhodococcus ruber Alcohol Dehydrogenase NADH Biomimetic Complex - Compound 4b | | Descriptor: | 1-{[4-(hydroxymethyl)phenyl]methyl}-1,4-dihydropyridine-3-carboxamide, CITRIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Wilson, L.A, Schenk, G, Guddat, L.W, Scott, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural Characterization of Enzymatic Interactions with Functional Nicotinamide Cofactor Biomimetics
Catalysts, 14, 2024
|
|
8UAK
 
 | | Crystal structure of the catalytic domain of human PKC alpha (D463N, V568I, S657E) in complex with Darovasertib (NVP-LXS196) at 2.82-A resolution | | Descriptor: | (6M)-3-amino-N-[3-(4-amino-4-methylpiperidin-1-yl)pyridin-2-yl]-6-[3-(trifluoromethyl)pyridin-2-yl]pyrazine-2-carboxamide, Protein kinase C alpha type | | Authors: | Romanowski, M.J, Lam, J, Visser, M. | | Deposit date: | 2023-09-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Discovery of Darovasertib (NVP-LXS196), a Pan-PKC Inhibitor for the Treatment of Metastatic Uveal Melanoma.
J.Med.Chem., 67, 2024
|
|
8UAI
 
 | |
8UAC
 
 | | CATHEPSIN L IN COMPLEX WITH AC1115 | | Descriptor: | Cathepsin L, N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-1H-indole-2-carboxamide | | Authors: | Chao, A, DuPrez, K.T, Han, F.Q. | | Deposit date: | 2023-09-20 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | CATHEPSIN L IN COMPLEX WITH AC1115
To be Published
|
|
8UAB
 
 | | SARS-CoV-2 main protease (Mpro) complex with AC1115 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-1H-indole-2-carboxamide | | Authors: | DuPrez, K.T, Chao, A, Han, F.Q. | | Deposit date: | 2023-09-20 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | SARS-CoV-2 main protease (Mpro) complex with AC1115
To Be Published
|
|
