2AIV
 
 | | Multiple conformations in the ligand-binding site of the yeast nuclear pore targeting domain of NUP116P | | Descriptor: | fragment of Nucleoporin NUP116/NSP116 | | Authors: | Robinson, M.A, Park, S, Sun, Z.-Y.J, Silver, P, Wagner, G, Hogle, J. | | Deposit date: | 2005-08-01 | | Release date: | 2005-08-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Multiple Conformations in the Ligand-binding Site of the Yeast Nuclear Pore-targeting Domain of Nup116p
J.Biol.Chem., 280, 2005
|
|
1R7X
 
 | | Glycosyltransferase B in complex with 3-amino-acceptor analog inhibitor | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MERCURY (II) ION, alpha-L-fucopyranose-(1-2)-hexyl 3-amino-3-deoxy-beta-D-galactopyranoside | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
2AXD
 
 | | solution structure of the theta subunit of escherichia coli DNA polymerase III in complex with the epsilon subunit | | Descriptor: | DNA polymerase III, theta subunit | | Authors: | Keniry, M.A, Park, A.Y, Owen, E.A, Hamdan, S.M, Pintacuda, G, Otting, G, Dixon, N.E. | | Deposit date: | 2005-09-05 | | Release date: | 2006-07-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the theta subunit of Escherichia coli DNA polymerase III in complex with the epsilon subunit
J.Bacteriol., 188, 2006
|
|
5X9X
 
 | |
1R7Y
 
 | | Glycosyltransferase A in complex with 3-amino-acceptor analog inhibitor and uridine diphosphate | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-N-acetylgalactosaminyltransferase, MANGANESE (II) ION, MERCURY (II) ION, ... | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
2B38
 
 | | Solution structure of kalata B8 | | Descriptor: | kalata B8 | | Authors: | Daly, N.L, Clark, R.J, Plan, M.R, Craik, D.J. | | Deposit date: | 2005-09-19 | | Release date: | 2006-01-31 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Kalata B8, a novel antiviral circular protein, exhibits conformational flexibility in the cystine knot motif
Biochem.J., 393, 2006
|
|
2MMC
 
 | | Nucleotide-free human ran gtpase | | Descriptor: | GTP-binding nuclear protein Ran | | Authors: | Bacot-Davis, V.R, Palmenberg, A.C. | | Deposit date: | 2014-03-13 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structure of Ran GTPase Determines C-terminal Tail Conformational Dynamics.
To be Published
|
|
5XA6
 
 | |
5X3Y
 
 | | Refined solution structure of musashi1 RBD2 | | Descriptor: | RNA-binding protein Musashi homolog 1 | | Authors: | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
8UEK
 
 | |
5IX9
 
 | | Cell surface anchoring domain | | Descriptor: | Antifreeze protein | | Authors: | Guo, S, Langelaan, D. | | Deposit date: | 2016-03-23 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
5XF0
 
 | |
5XND
 
 | |
1TQZ
 
 | |
5XHT
 
 | | The PHD finger of human Kiaa1045 protein | | Descriptor: | PHD finger protein 24, ZINC ION | | Authors: | Miyamoto, K. | | Deposit date: | 2017-04-24 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PHD finger from the human KIAA1045 protein
Protein Sci., 27, 2018
|
|
5JTN
 
 | | The structure of chaperone SecB in complex with unstructured proPhoA binding site c | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5JTL
 
 | | The structure of chaperone SecB in complex with unstructured proPhoA | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
6L6V
 
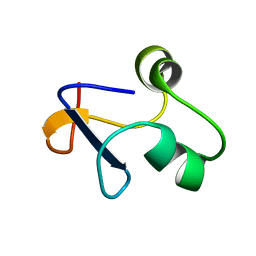 | | SPO1 Gp44 N-terminal region (1-55) | | Descriptor: | E3 protein | | Authors: | Liu, B, Wang, Z. | | Deposit date: | 2019-10-29 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Bacteriophage DNA Mimic Protein Employs a Non-specific Strategy to Inhibit the Bacterial RNA Polymerase.
Front Microbiol, 12, 2021
|
|
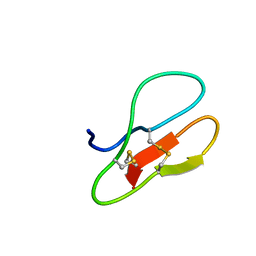
5X38
 
 | |
5X35
 
 | |
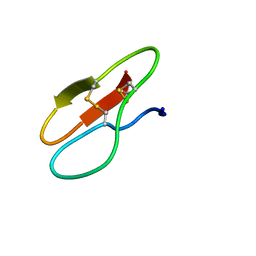
5X36
 
 | |
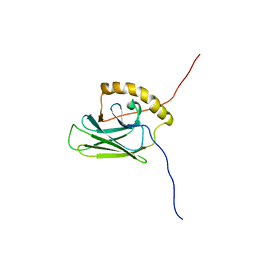
5XI9
 
 | |
5X37
 
 | |
5X3C
 
 | |
5XIR
 
 | |
