2TLD
 
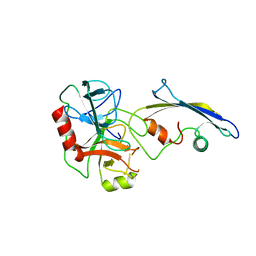 | | CRYSTAL STRUCTURE OF AN ENGINEERED SUBTILISIN INHIBITOR COMPLEXED WITH BOVINE TRYPSIN | | Descriptor: | STREPTOMYCES SUBTILISIN INHIBITOR (SSI), TRYPSIN | | Authors: | Mitsui, Y, Takeuchi, Y, Nonaka, T, Nakamura, K.T. | | Deposit date: | 1991-09-16 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an engineered subtilisin inhibitor complexed with bovine trypsin.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
1HDD
 
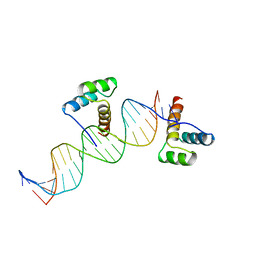 | | CRYSTAL STRUCTURE OF AN ENGRAILED HOMEODOMAIN-DNA COMPLEX AT 2.8 ANGSTROMS RESOLUTION: A FRAMEWORK FOR UNDERSTANDING HOMEODOMAIN-DNA INTERACTIONS | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*GP*GP*TP*AP*AP*TP*TP*AP*CP*AP*TP*GP*G P*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*TP*AP*C P*CP*TP*AP*A)-3'), PROTEIN (ENGRAILED HOMEODOMAIN) | | Authors: | Kissinger, C.R, Liu, B, Martin-Blanco, E, Kornberg, T.B, Pabo, C.O. | | Deposit date: | 1991-09-16 | | Release date: | 1992-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions.
Cell(Cambridge,Mass.), 63, 1990
|
|
1PK2
 
 | |
1CCD
 
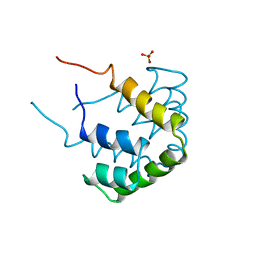 | | REFINED STRUCTURE OF RAT CLARA CELL 17 KDA PROTEIN AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | CLARA CELL 17 kD PROTEIN, SULFATE ION | | Authors: | Umland, T.C, Swaminathan, S, Furey, W, Singh, G, Pletcher, J, Sax, M. | | Deposit date: | 1991-09-17 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Refined structure of rat Clara cell 17 kDa protein at 3.0 A resolution.
J.Mol.Biol., 224, 1992
|
|
1D49
 
 | | THE STRUCTURE OF A B-DNA DECAMER WITH A CENTRAL T-A STEP: C-G-A-T-T-A-A-T-C-G | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*TP*AP*AP*TP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Grzeskowiak, K, Yanagi, K, Dickerson, R.E. | | Deposit date: | 1991-09-17 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a B-DNA decamer with a central T-A step: C-G-A-T-T-A-A-T-C-G.
J.Mol.Biol., 225, 1992
|
|
1GBT
 
 | |
1HIO
 
 | | HISTONE OCTAMER (CHICKEN), CHROMOSOMAL PROTEIN, ALPHA CARBONS ONLY | | Descriptor: | HISTONE H2A, HISTONE H2B, HISTONE H3, ... | | Authors: | Arents, G, Moudrianakis, E.N. | | Deposit date: | 1991-09-19 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1BBI
 
 | |
2BBI
 
 | |
3TMS
 
 | |
1L72
 
 | |
1MSB
 
 | |
1L73
 
 | |
1L70
 
 | |
1L75
 
 | |
1L65
 
 | |
1L67
 
 | |
1L74
 
 | |
1L69
 
 | |
1L68
 
 | |
8RNT
 
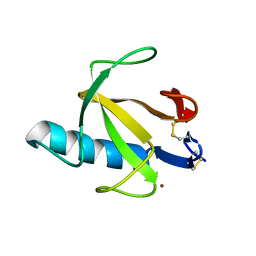 | | STRUCTURE OF RIBONUCLEASE T1 COMPLEXED WITH ZINC(II) AT 1.8 ANGSTROMS RESOLUTION: A ZN2+.6H2O.CARBOXYLATE CLATHRATE | | Descriptor: | RIBONUCLEASE T1, ZINC ION | | Authors: | Ding, J, Choe, H.-W, Granzin, J, Saenger, W. | | Deposit date: | 1991-09-23 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of ribonuclease T1 complexed with zinc(II) at 1.8 A resolution: a Zn2+.6H2O.carboxylate clathrate.
Acta Crystallogr.,Sect.B, 48, 1992
|
|
1L64
 
 | |
1L71
 
 | |
1L76
 
 | |
1L66
 
 | |
