8GXS
 
 | | PIC-Mediator in complex with +1 nucleosome (T40N) in H-binding state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wang, X, Liu, W, Ren, Y, Qu, X, Li, J, Yin, X. | | Deposit date: | 2022-09-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structures of +1 nucleosome-bound PIC-Mediator complex.
Science, 378, 2022
|
|
9K3V
 
 | | Human RNA Polymerase III de novo transcribing complex 4 (TC4) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (96-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-20 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action.
Nature, 2025
|
|
9K3U
 
 | | Human RNA Polymerase III de novo transcribing complex 5 (TC5) | | Descriptor: | DNA (97-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-20 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action.
Nature, 2025
|
|
9K3B
 
 | | Human RNA Polymerase III de novo transcribing complex 10 overall (TC10-overall) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (102-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-18 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action.
Nature, 2025
|
|
8G60
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (CR state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G61
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (AC state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G5Z
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G6J
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state 2) | | Descriptor: | (3R,6R,9S,12S,15S,18S,20R,24aR)-6-[(2S)-butan-2-yl]-3,12-bis[(1R)-1-hydroxy-2-methylpropyl]-8,9,11,17,18-pentamethyl-15-[(2S)-2-methylbutyl]hexadecahydropyrido[1,2-a][1,4,7,10,13,16,19]heptaazacyclohenicosine-1,4,7,10,13,16,19(21H)-heptone, (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 1,4-DIAMINOBUTANE, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-15 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G5Y
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (IC state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8GLP
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (Consensus LSU focused refined structure) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Watson, Z.L, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-03-22 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (1.67 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8UOQ
 
 | | Composite map of PIC_delta_TFIIK form2 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2023-10-20 | | Release date: | 2024-10-23 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Transcription start site scanning requires the fungi-specific hydrophobic loop of Tfb3.
Nucleic Acids Res., 52, 2024
|
|
8UOT
 
 | | Composite map of PICdeltaTFIIK form1 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2023-10-20 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Transcription start site scanning requires the fungi-specific hydrophobic loop of Tfb3.
Nucleic Acids Res., 52, 2024
|
|
9K36
 
 | | Human RNA Polymerase III de novo transcribing complex 12 (TC12) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (54-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-18 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action.
Nature, 2025
|
|
9K2G
 
 | | Human RNA Polymerase III de novo transcribing complex 13 (TC13) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (104-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-17 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action.
Nature, 2025
|
|
9K39
 
 | | Human RNA Polymerase III de novo transcribing complex 8 (TC8) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (50-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-18 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action.
Nature, 2025
|
|
9K38
 
 | | Human RNA Polymerase III de novo transcribing complex 6 (TC6) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (51-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-18 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action.
Nature, 2025
|
|
6EZ4
 
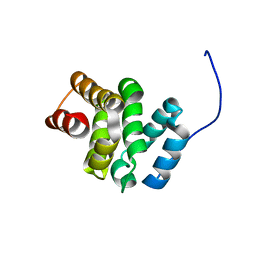 | | NMR structure of the C-terminal domain of the human RPAP3 protein | | Descriptor: | RNA polymerase II-associated protein 3 | | Authors: | Fabre, P, Chagot, M.E, Bragantini, B, Manival, X, Quinternet, M. | | Deposit date: | 2017-11-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | The RPAP3-Cterminal domain identifies R2TP-like quaternary chaperones.
Nat Commun, 9, 2018
|
|
7Z1L
 
 | | Structure of yeast RNA Polymerase III Pre-Termination Complex (PTC) | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1O
 
 | | Structure of yeast RNA Polymerase III PTC + NTPs | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1M
 
 | | Structure of yeast RNA Polymerase III Elongation Complex (EC) | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1N
 
 | | Structure of yeast RNA Polymerase III Delta C53-C37-C11 | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
8RN9
 
 | |
8RN2
 
 | | Monomeric apo-influenza B polymerase, encapsidase conformation | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Nat Commun, 15, 2024
|
|
8RN6
 
 | |
6T0R
 
 | | Bat Influenza A polymerase product dissociation complex using 44-mer vRNA template with mutated oligo(U) sequence | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit, ... | | Authors: | Wandzik, J.M, Kouba, T, Cusack, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-15 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 181, 2020
|
|
