1H8B
 
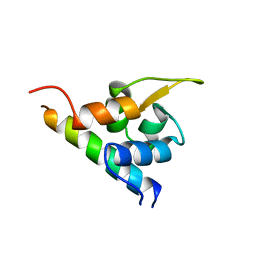 | | EF-hands 3,4 from alpha-actinin / Z-repeat 7 from titin | | Descriptor: | ALPHA-ACTININ 2, SKELETAL MUSCLE ISOFORM, TITIN | | Authors: | Atkinson, R.A, Joseph, C, Kelly, G, Muskett, F.W, Frenkiel, T.A, Nietlispach, D, Pastore, A. | | Deposit date: | 2001-02-01 | | Release date: | 2001-08-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ca2+-Independent Binding of an EF-Hand Domain to a Novel Motif in the Alpha-Actinin-Titin Complex
Nat.Struct.Biol., 8, 2001
|
|
5N4C
 
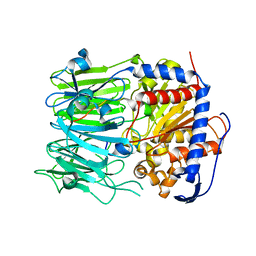 | | Prolyl oligopeptidase B from Galerina marginata bound to 35mer hydrolysis and macrocyclization substrate - S577A mutant | | Descriptor: | Alpha-amanitin proprotein, GLYCEROL, Prolyl oligopeptidase | | Authors: | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
6E0X
 
 | |
7MFW
 
 | |
5MFC
 
 | | Designed armadillo repeat protein YIIIM5AII in complex with (KR)4-GFP | | Descriptor: | (KR)4-Green fluorescent protein,Green fluorescent protein, ACETATE ION, YIIIM5AII | | Authors: | Hansen, S, Kiefer, J, Madhurantakam, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-07-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of designed armadillo repeat proteins binding to peptides fused to globular domains.
Protein Sci., 26, 2017
|
|
2WAH
 
 | | Crystal Structure of an IgG1 Fc Glycoform (Man9GlcNAc2) | | Descriptor: | IG GAMMA-1 CHAIN C REGION, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Crispin, M, Bowden, T.A, Coles, C.H, Harlos, K, Aricescu, A.R, Harvey, D.J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2009-02-06 | | Release date: | 2009-03-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Carbohydrate and Domain Architecture of an Immature Antibody Glycoform Exhibiting Enhanced Effector Functions
J.Mol.Biol., 387, 2009
|
|
9H6X
 
 | | Cryo-EM structure of lipid-bound human myoferlin (25 mol% DOPS/5 mol% PI(4,5)P2 nanodisc) | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, CALCIUM ION, Myoferlin | | Authors: | Cretu, C, Moser, T, Preobraschenski, J. | | Deposit date: | 2024-10-25 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural insights into lipid membrane binding by human ferlins.
Embo J., 2025
|
|
3LHS
 
 | | Open Conformation of HtsA Complexed with Staphyloferrin A | | Descriptor: | (2R)-2-(2-{[(1R)-1-carboxy-4-{[(3S)-3,4-dicarboxy-3-hydroxybutanoyl]amino}butyl]amino}-2-oxoethyl)-2-hydroxybutanedioic acid, FE (III) ION, Ferrichrome ABC transporter lipoprotein, ... | | Authors: | Grigg, J.C, Murphy, M.E.P. | | Deposit date: | 2010-01-23 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Staphylococcus aureus siderophore receptor HtsA undergoes localized conformational changes to enclose staphyloferrin A in an arginine-rich binding pocket.
J.Biol.Chem., 285, 2010
|
|
6WR5
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J35 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*AP*TP*GP*AP*CP*GP*AP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-04-29 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.063 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WR7
 
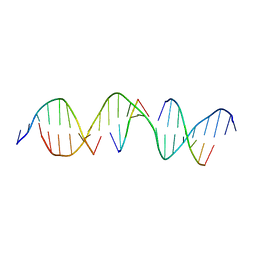 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J33 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*TP*TP*GP*AP*CP*AP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), DNA (5'-D(P*AP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-04-29 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WRB
 
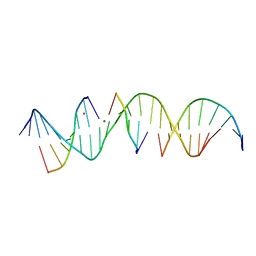 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J5 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*CP*GP*AP*CP*GP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-04-29 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WRI
 
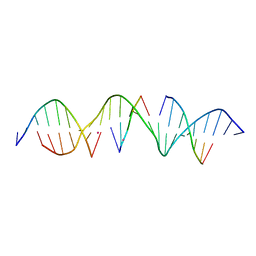 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J28 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*AP*TP*GP*AP*CP*TP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-04-29 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.057 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSU
 
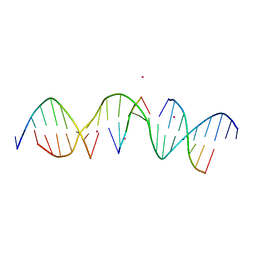 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J19 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*AP*GP*AP*CP*TP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6E2J
 
 | | Crystal structure of the heterocomplex between human keratin 1 coil 1B containing S233L mutation and wild-type human keratin 10 coil 1B | | Descriptor: | Keratin, type I cytoskeletal 10, type II cytoskeletal 1, ... | | Authors: | Eldirany, S.A, Lomakin, I.B, Bunick, C.G. | | Deposit date: | 2018-07-11 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.386 Å) | | Cite: | Human keratin 1/10-1B tetramer structures reveal a knob-pocket mechanism in intermediate filament assembly.
Embo J., 38, 2019
|
|
6WSV
 
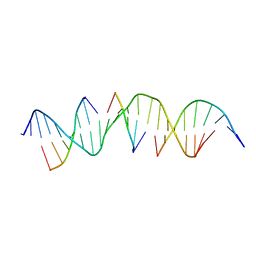 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J20 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*GP*AP*CP*AP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSS
 
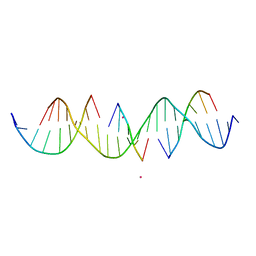 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J15 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*GP*AP*CP*CP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSP
 
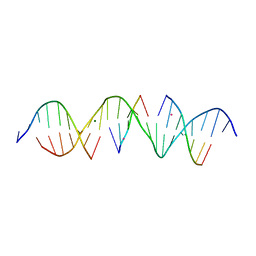 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J9 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*GP*AP*CP*TP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.049 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSR
 
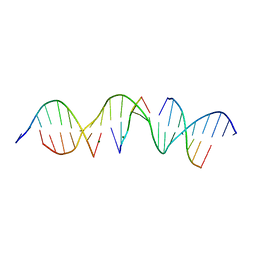 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J10 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*AP*GP*AP*CP*CP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), DNA (5'-D(P*CP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSZ
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J24 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*GP*AP*CP*TP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.053 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSW
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J21 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*AP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*TP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
2XX0
 
 | | STRUCTURE OF THE N90S-H254F MUTANT OF NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Leferink, N.G.H, Han, C, Heyes, D.J, Rigby, S.E.J, Hough, M.A, Eady, R.R, Scrutton, N.S, Hasnain, S.S. | | Deposit date: | 2010-11-07 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Proton-Coupled Electron Transfer in the Catalytic Cycle of Alcaligenes Xylosoxidans Copper-Dependent Nitrite Reductase.
Biochemistry, 50, 2011
|
|
6WT0
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J25 immobile Holliday junction | | Descriptor: | COBALT (II) ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*AP*GP*AP*CP*CP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WT1
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J29 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*AP*AP*GP*AP*CP*TP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), DNA (5'-D(P*AP*GP*TP*CP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
8EC6
 
 | | Cryo-EM structure of the Glutaminase C core filament (fGAC) | | Descriptor: | Isoform 2 of Glutaminase kidney isoform, mitochondrial, PHOSPHATE ION | | Authors: | Ambrosio, A.L, Dias, S.M, Quesnay, J.E, Portugal, R.V, Cassago, A, van Heel, M.G, Islam, Z, Rodrigues, C.T. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism of glutaminase activation through filamentation and the role of filaments in mitophagy protection.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2BOD
 
 | | Catalytic domain of endo-1,4-glucanase Cel6A from Thermobifida fusca in complex with methyl cellobiosyl-4-thio-beta-cellobioside | | Descriptor: | ENDOGLUCANASE E-2, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-methyl beta-D-glucopyranoside | | Authors: | Larsson, A.M, Bergfors, T, Dultz, E, Irwin, D.C, Roos, A, Driguez, H, Wilson, D.B, Jones, T.A. | | Deposit date: | 2005-04-10 | | Release date: | 2005-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Thermobifida Fusca Endoglucanase Cel6A in Complex with Substrate and Inhibitor: The Role of Tyrosine Y73 in Substrate Ring Distortion.
Biochemistry, 44, 2005
|
|
