8ARX
 
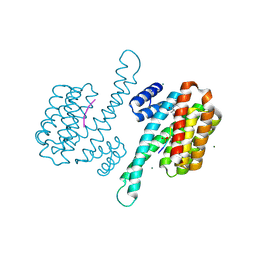 | | Small molecular stabilizer for ERalpha and 14-3-3sigma (1074378) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[[1-[1-(4-chloranylphenoxy)cyclopropyl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8A98
 
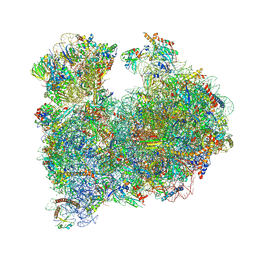 | | CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME : snoRNA MUTANT | | Descriptor: | 40S ribosomal protein S12, 40S ribosomal protein S14, 40S ribosomal protein S19-like protein, ... | | Authors: | Rajan, K.S, Yonath, A, Bashan, A. | | Deposit date: | 2022-06-28 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural and mechanistic insights into the function of Leishmania ribosome lacking a single pseudouridine modification.
Cell Rep, 43, 2024
|
|
8QK9
 
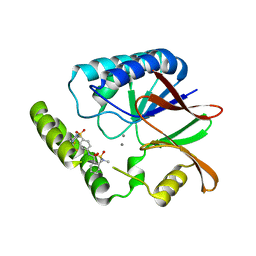 | | Structure of E. coli LpxH in complex with JEDI-1444 | | Descriptor: | 2-[methyl(methylsulfonyl)amino]-~{N}-[4-[4-[3-(trifluoromethyl)phenyl]piperazin-1-yl]sulfonylphenyl]benzamide, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6P91
 
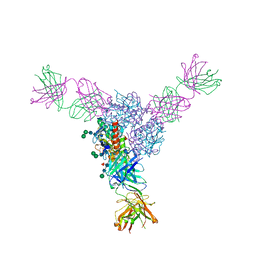 | | Structure of Lassa virus glycoprotein bound to Fab 18.5C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 18.5C Antibody heavy chain, ... | | Authors: | Saphire, E.O, Hastie, K.M. | | Deposit date: | 2019-06-09 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Convergent Structures Illuminate Features for Germline Antibody Binding and Pan-Lassa Virus Neutralization.
Cell, 178, 2019
|
|
7RRX
 
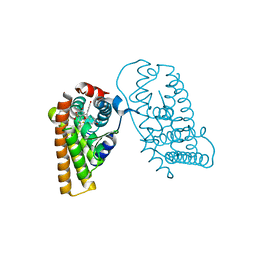 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-19 | | Descriptor: | (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-N-{4-[2-(piperidin-1-yl)ethoxy]phenyl}-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, CHLORIDE ION, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RS8
 
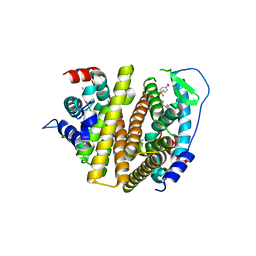 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-16 | | Descriptor: | (1R,2S,4R)-5-(4-hydroxyphenyl)-N-(4-methoxyphenyl)-6-{4-[2-(piperidin-1-yl)ethoxy]phenyl}-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RS7
 
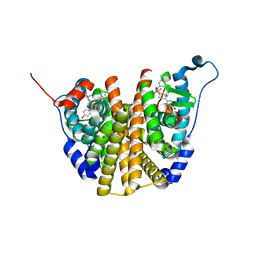 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-30 | | Descriptor: | (1S,2R,4S,5S,6S)-N,5,6-tris(4-hydroxyphenyl)-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]heptane-2-sulfonamide, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7U5E
 
 | | I-F3b Cascade-TniQ partial R-loop complex | | Descriptor: | Cas6, Cas7, Cas8/5, ... | | Authors: | Park, J.U, Mehrotra, E, Kellogg, E.H. | | Deposit date: | 2022-03-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Multiple adaptations underly co-option of a CRISPR surveillance complex for RNA-guided DNA transposition.
Mol.Cell, 83, 2023
|
|
7AA1
 
 | | Structural comparison of cellular retinoic acid binding proteins I and II in the presence and absence of natural and synthetic ligands | | Descriptor: | 4-[2-(5,5,8,8-tetramethyl-6,7-dihydroquinoxalin-2-yl)ethynyl]benzoic acid, Cellular retinoic acid-binding protein 2 | | Authors: | Tomlinson, C.W.E, Cornish, K.A.S, Pohl, E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure-functional relationship of cellular retinoic acid-binding proteins I and II interacting with natural and synthetic ligands.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7U5D
 
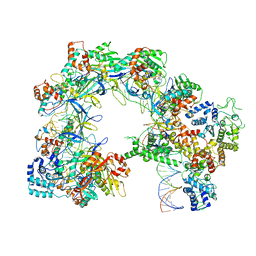 | | I-F3b Cascade-TniQ full R-loop complex | | Descriptor: | Cas6, Cas7, Cas8/5, ... | | Authors: | Park, J.U, Mehrotra, E, Kellogg, E.H. | | Deposit date: | 2022-03-02 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Multiple adaptations underly co-option of a CRISPR surveillance complex for RNA-guided DNA transposition.
Mol.Cell, 83, 2023
|
|
6XKE
 
 | |
8AKU
 
 | | 250 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
8AKY
 
 | | 290 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
7A6Q
 
 | | Crystal structure of human aldehyde dehydrogenase 1A3 in complex with selective NR6 inhibitor compound | | Descriptor: | (3-oxidanylidene-3-sodiooxy-propanoyl)oxysodium, 3-(2-phenylimidazo[1,2-a]pyridin-6-yl)benzenecarbonitrile, Aldehyde dehydrogenase family 1 member A3, ... | | Authors: | Gelardi, E.L.M, Garavaglia, S. | | Deposit date: | 2020-08-26 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A Selective Competitive Inhibitor of Aldehyde Dehydrogenase 1A3 Hinders Cancer Cell Growth, Invasiveness and Stemness In Vitro.
Cancers (Basel), 13, 2021
|
|
8AKZ
 
 | | 320 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
8AKQ
 
 | | 180 A SynPspA rod after incubation with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
6XL7
 
 | |
8AKR
 
 | | 200 A SynPspA rod after incubation with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
7RGB
 
 | | O2-, PLP-dependent desaturase Plu4 product-bound enzyme | | Descriptor: | (2Z,4E)-5-carbamimidamido-2-iminopent-4-enoic acid, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Hoffarth, E.R, Ryan, K.S. | | Deposit date: | 2021-07-14 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A shared mechanistic pathway for pyridoxal phosphate-dependent arginine oxidases.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8AKS
 
 | | 215 A SynPspA rod after incubation with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
7RF9
 
 | | O2-, PLP-dependent desaturase Plu4 intermediate-bound enzyme | | Descriptor: | (2E)-5-carbamimidamido-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}pentanoic acid, 1,2-ETHANEDIOL, 2-(2-ETHOXYETHOXY)ETHANOL, ... | | Authors: | Hoffarth, E.R, Ryan, K.S. | | Deposit date: | 2021-07-13 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | A shared mechanistic pathway for pyridoxal phosphate-dependent arginine oxidases.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8AKT
 
 | | 235 A SynPspA rod after incubation with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
8AL0
 
 | | 365 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
8AKX
 
 | | 305 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
8AKV
 
 | | 270 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
