1ZFA
 
 | | GGA Duplex A-DNA | | Descriptor: | 5'-D(*CP*CP*TP*CP*CP*GP*GP*AP*GP*G)-3', CALCIUM ION, SODIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZHM
 
 | | Crystal Structure of the Catalytic Domain of the Coagulation Factor XIa in Complex with Benzamidine (S434A-T475A-K437 Mutant) | | Descriptor: | BENZAMIDINE, GLUTATHIONE, coagulation factor XI | | Authors: | Jin, L, Pandey, P, Babine, R.E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2005-04-26 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Mutation of surface residues to promote crystallization of activated factor XI as a complex with benzamidine: an essential step for the iterative structure-based design of factor XI inhibitors.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4N16
 
 | | Structure of cholate bound to human carbonic anhydrase II | | Descriptor: | CHOLIC ACID, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Boone, C.D, McKenna, R. | | Deposit date: | 2013-10-03 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural elucidation of the hormonal inhibition mechanism of the bile acid cholate on human carbonic anhydrase II.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4I71
 
 | | Crystal structure of the Trypanosoma brucei Inosine-Adenosine-Guanosine nucleoside hydrolase in complex with a trypanocidal compound | | Descriptor: | (2R,3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-2-(hydroxymethyl)pyrrolidine-3,4-diol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Giannese, F, Degano, M. | | Deposit date: | 2012-11-30 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structures of purine nucleosidase from Trypanosoma brucei bound to isozyme-specific trypanocidals and a novel metalorganic inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1YYM
 
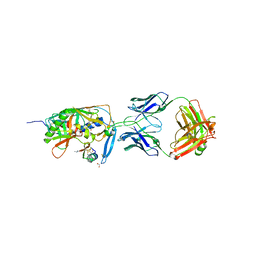 | | crystal structure of F23, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 gp120 envelope glycoprotein and anti-HIV-1 antibody 17b | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Exterior membrane glycoprotein(GP120),Exterior membrane glycoprotein(GP120),Exterior membrane glycoprotein(GP120), ... | | Authors: | Huang, C.C, Stricher, F, Martin, L, Decker, J.M, Majeed, S, Barthe, P, Hendrickson, W.A, Robinson, J, Roumestand, C, Sodroski, J, Wyatt, R, Shaw, G.M, Vita, C, Kwong, P.D. | | Deposit date: | 2005-02-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth.
Structure, 13, 2005
|
|
4N8Z
 
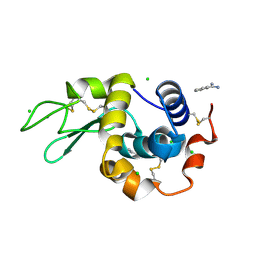 | | In situ lysozyme crystallized on a MiTeGen micromesh with benzamidine ligand | | Descriptor: | BENZAMIDINE, CHLORIDE ION, Lysozyme C, ... | | Authors: | Yin, X, Scalia, A, Leroy, L, Cuttitta, C.M, Polizzo, G.M, Ericson, D.L, Roessler, C.G, Campos, O, Agarwal, R, Allaire, M, Orville, A.M, Jackimowicz, R, Ma, M.Y, Sweet, R.M, Soares, A.S. | | Deposit date: | 2013-10-18 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Hitting the target: fragment screening with acoustic in situ co-crystallization of proteins plus fragment libraries on pin-mounted data-collection micromeshes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WV3
 
 | | Crystal structure of the catalytic domain of MMP-13 complexed with N-(3-methoxybenzyl)-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Collagenase 3, ... | | Authors: | Oki, H, Tanaka, Y. | | Deposit date: | 2014-05-12 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Thieno[2,3-d]pyrimidine-2-carboxamides bearing a carboxybenzene group at 5-position: highly potent, selective, and orally available MMP-13 inhibitors interacting with the S1′′ binding site.
Bioorg.Med.Chem., 22, 2014
|
|
1ZF9
 
 | | GGG Duplex A-DNA | | Descriptor: | 5'-D(*CP*CP*CP*CP*CP*GP*GP*GP*GP*G)-3', SODIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3W7A
 
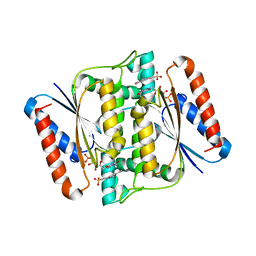 | | Crystal Structure of azoreductase AzrC fin complex with sulfone-modified azo dye Acid Red 88 | | Descriptor: | 4-[(E)-(2-hydroxynaphthalen-1-yl)diazenyl]naphthalene-1-sulfonic acid, CALCIUM ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Yu, J, Ogata, D, Ooi, T, Yao, M. | | Deposit date: | 2013-02-27 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of AzrA and of AzrC complexed with substrate or inhibitor: insight into substrate specificity and catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LU9
 
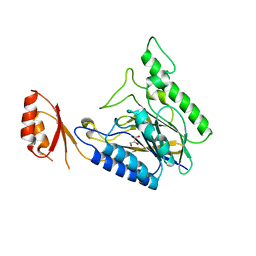 | | Crystal structure of E.coli SbcD at 2.5 angstrom resolution | | Descriptor: | Exonuclease subunit SbcD, GLYCEROL | | Authors: | Liu, S, Tian, L.F, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-07-25 | | Release date: | 2014-08-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for DNA recognition and nuclease processing by the Mre11 homologue SbcD in double-strand breaks repair.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5QDU
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000466a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, ~{N},~{N},5,6-tetramethylthieno[2,3-d]pyrimidin-4-amine | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.672 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
1L4I
 
 | | Crystal Structure of the Periplasmic Chaperone SfaE | | Descriptor: | SfaE PROTEIN | | Authors: | Knight, S.D, Choudhury, D, Hultgren, S, Pinkner, J, Stojanoff, V, Thompson, A. | | Deposit date: | 2002-03-05 | | Release date: | 2002-06-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the S pilus periplasmic chaperone SfaE at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1UF5
 
 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase complexed with N-carbamyl-D-methionine | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYLSULFANYL-2-UREIDO-BUTYRIC ACID, N-carbamyl-D-amino acid amidohydrolase | | Authors: | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | Deposit date: | 2003-05-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of C171A/V236A mutant of N-carbamyl-D-amino acid amidohydrolase
To be published
|
|
4L19
 
 | | Matrix metalloproteinase-13 complexed with selective inhibitor compound Q1 | | Descriptor: | 2-[(4-methylbenzyl)sulfanyl]-3,5,6,7-tetrahydro-4H-cyclopenta[d]pyrimidin-4-one, CALCIUM ION, Collagenase 3, ... | | Authors: | Minond, D, Spicer, T.P, Jiang, J, Taylor, A.B, Hart, P.J, Roush, W.R, Fields, G.B, Hodder, P.S. | | Deposit date: | 2013-06-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Characterization of selective exosite-binding inhibitors of matrix metalloproteinase 13 that prevent articular cartilage degradation in vitro.
J.Med.Chem., 57, 2014
|
|
1KZA
 
 | | Complex of MBP-C and Man-a13-Man | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN C, ... | | Authors: | Ng, K.K, Kolatkar, A.R, Park-Snyder, S, Feinberg, H, Clark, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Orientation of bound ligands in mannose-binding proteins. Implications for multivalent ligand recognition.
J.Biol.Chem., 277, 2002
|
|
3OPF
 
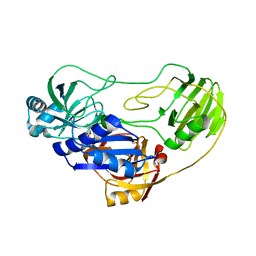 | | Crystal structure of TTHA0988 in space group P212121 | | Descriptor: | GLYCEROL, Putative uncharacterized protein TTHA0988, SULFATE ION | | Authors: | Jacques, D.A, Kuramitsu, S, Yokoyama, S, Trewhella, J, Guss, J.M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-01 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of TTHA0988 from Thermus thermophilus, a KipI-KipA homologue incorrectly annotated as an allophanate hydrolase
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2JCQ
 
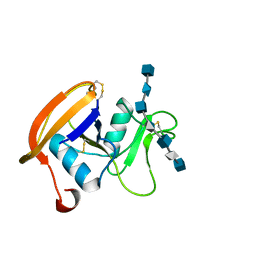 | | The hyaluronan binding domain of murine CD44 in a Type A complex with an HA 8-mer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD44 ANTIGEN, GLYCEROL | | Authors: | Banerji, S, Wright, A.J, Noble, M.E.M, Mahoney, D.J, Campbell, I.D, Day, A.J, Jackson, D.G. | | Deposit date: | 2007-01-03 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structures of the Cd44-Hyaluronan Complex Provide Insight Into a Fundamental Carbohydrate-Protein Interaction.
Nat.Struct.Mol.Biol., 14, 2008
|
|
1G0W
 
 | |
3F64
 
 | | F17a-G lectin domain with bound GlcNAc(beta1-O)paranitrophenyl ligand | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, F17a-G | | Authors: | Buts, L, De Boer, A, Olsson, J.D.M, Jonckheere, W, De Kerpel, M, De Genst, E, Guerardel, Y, Willaert, R, Wyns, L, Wuhrer, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-11-05 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli.
Biology (Basel), 2, 2013
|
|
1Y8J
 
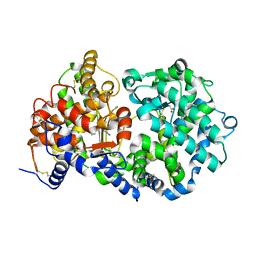 | | Crystal Structure of human NEP complexed with an imidazo[4,5-c]pyridine inhibitor | | Descriptor: | 2-[(1S)-1-BENZYL-2-SULFANYLETHYL]-1H-IMIDAZO[4,5-C]PYRIDIN-5-IUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Sahli, S, Frank, B, Schweizer, W.B, Diederich, F, Blum-Kaelin, D, Aebi, J.D, Bohm, H.J, Oefner, C, Dale, G.E. | | Deposit date: | 2004-12-13 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Second-Generation Inhibitors for the Metalloprotease Neprilysin Based on Bicyclic Heteroaromatic Scaffolds: Synthesis, Biological Activity, and X-ray Crystal Structure Analysis
HELV.CHIM.ACTA, 88, 2005
|
|
3FBA
 
 | | Crystal structure of 2C-methyl-D-erythritol 2,4-clycodiphosphate synthase complexed with ligand | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, GERANYL DIPHOSPHATE, GLYCEROL, ... | | Authors: | Hunter, W.N, Ramsden, N.L. | | Deposit date: | 2008-11-19 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A structure-based approach to ligand discovery for 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase: a target for antimicrobial therapy
J.Med.Chem., 52, 2009
|
|
4IYR
 
 | | Crystal structure of full-length caspase-6 zymogen | | Descriptor: | Caspase-6 | | Authors: | Cao, Q, Wang, X.-J, Li, L.-F, Su, X.-D. | | Deposit date: | 2013-01-29 | | Release date: | 2014-01-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | The regulatory mechanism of the caspase 6 pro-domain revealed by crystal structure and biochemical assays.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1L6J
 
 | | Crystal structure of human matrix metalloproteinase MMP9 (gelatinase B). | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-9, ZINC ION | | Authors: | Elkins, P.A, Ho, Y.S, Smith, W.W, Janson, C.A, D'Alessio, K.J, McQueney, M.S, Cummings, M.D, Romanic, A.M. | | Deposit date: | 2002-03-11 | | Release date: | 2002-07-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the C-terminally truncated human ProMMP9, a gelatin-binding matrix metalloproteinase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1CS2
 
 | | NMR STRUCTURES OF B-DNA D(CTACTGCTTTAG).D(CTAAAGCAGTAG) | | Descriptor: | 5'-d(*CP*TP*AP*AP*AP*GP*CP*AP*GP*TP*AP*G)-3', 5'-d(*CP*TP*AP*CP*TP*GP*CP*TP*TP*TP*AP*G)-3' | | Authors: | Leporc, S, Mauffret, O, Tevanian, G, Lescot, E, Monnot, M, Fermandjian, S. | | Deposit date: | 1999-08-16 | | Release date: | 1999-08-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR and molecular modelling analysis of d(CTACTGCTTTAG). d(CTAAAGCAGTAG) reveals that the particular behaviour of TpA steps is related to edge-to-edge contacts of their base-pairs in the major groove
Nucleic Acids Res., 27, 1999
|
|
1UHY
 
 | | Crystal structure of d(GCGATAGC): the base-intercalated duplex | | Descriptor: | 5'-D(*GP*(CBR)P*GP*AP*TP*AP*GP*C)-3', CHLORIDE ION, COBALT HEXAMMINE(III), ... | | Authors: | Kondo, J, Umeda, S.I, Fujita, K, Sunami, T, Takenaka, A. | | Deposit date: | 2003-07-13 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray analyses of d(GCGAXAGC) containing G and T at X: the base-intercalated duplex is still stable even in point mutants at the fifth residue.
J.Synchrotron Radiat., 11, 2004
|
|
