1PEX
 
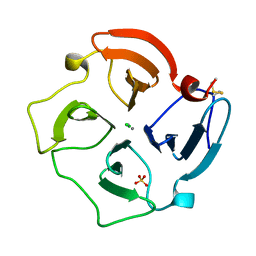 | | COLLAGENASE-3 (MMP-13) C-TERMINAL HEMOPEXIN-LIKE DOMAIN | | Descriptor: | CALCIUM ION, CHLORIDE ION, COLLAGENASE-3, ... | | Authors: | Gomis-Ruth, F.X, Gohlke, U, Betz, M, Knauper, V, Murphy, G, Lopez-Otin, C, Bode, W. | | Deposit date: | 1996-05-24 | | Release date: | 1996-12-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The helping hand of collagenase-3 (MMP-13): 2.7 A crystal structure of its C-terminal haemopexin-like domain.
J.Mol.Biol., 264, 1996
|
|
3SU3
 
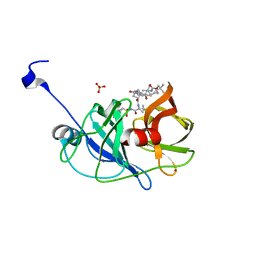 | | Crystal structure of NS3/4A protease in complex with vaniprevir | | Descriptor: | (5R,7S,10S)-10-tert-butyl-N-{(1R,2R)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethylcyclopropyl}-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19-dodecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosine-7(3H)-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SUG
 
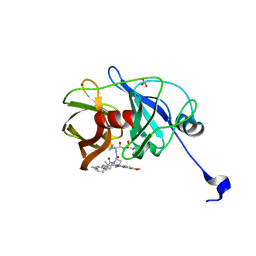 | | Crystal structure of NS3/4A protease variant A156T in complex with MK-5172 | | Descriptor: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-di oxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadec ino[11,12-b]quinoxaline-8-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SU6
 
 | | Crystal structure of NS3/4A protease variant A156T in complex with vaniprevir | | Descriptor: | (5R,7S,10S)-10-tert-butyl-N-{(1R,2R)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethylcyclopropyl}-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19-dodecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosine-7(3H)-carboxamide, GLYCEROL, NS3 protease, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SU1
 
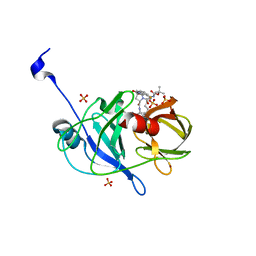 | | Crystal structure of NS3/4A protease variant D168A in complex with danoprevir | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, Genome polyprotein, SULFATE ION, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
4PTD
 
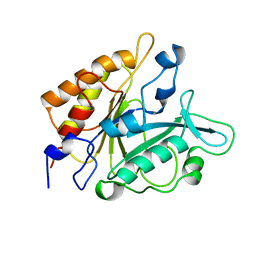 | | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C MUTANT D274N | | Descriptor: | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Heinz, D.W. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the roles of active site residues in phosphatidylinositol-specific phospholipase C from Bacillus cereus by site-directed mutagenesis.
Biochemistry, 36, 1997
|
|
1KFZ
 
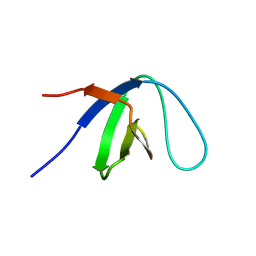 | | Solution Structure of C-terminal Sem-5 SH3 Domain (Ensemble of 16 Structures) | | Descriptor: | SEX MUSCLE ABNORMAL PROTEIN 5 | | Authors: | Ferreon, J.C, Volk, D.E, Luxon, B.A, Gorenstein, D, Hilser, V.J. | | Deposit date: | 2001-11-24 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Dynamics and Thermodynamics of the Native State Ensemble
of Sem-5 C-Terminal SH3 Domain
Biochemistry, 42, 2003
|
|
5PZN
 
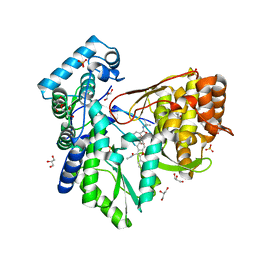 | |
5PZO
 
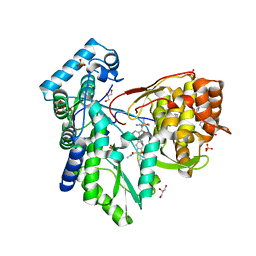 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE C316N IN COMPLEX WITH 2-(4-FLUOROPHENYL)-N-METHYL-5-[3-({2-METHYL-1-OXO-1-[(1,3,4-THIADIAZOL-2-YL)AMINO]PROPAN-2-YL}CARBAMOYL)PHENYL]-1-BENZOFURAN-3-CARBOXAMIDE | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, 2-(4-fluorophenyl)-N-methyl-5-[3-({2-methyl-1-oxo-1-[(1,3,4-thiadiazol-2-yl)amino]propan-2-yl}carbamoyl)phenyl]-1-benzofuran-3-carboxamide, GLYCEROL, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-27 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a Hepatitis C Virus NS5B Replicase Palm Site Allosteric Inhibitor (BMS-929075) Advanced to Phase 1 Clinical Studies.
J. Med. Chem., 60, 2017
|
|
1ALC
 
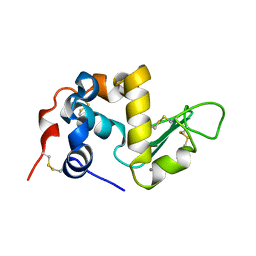 | |
1ZI0
 
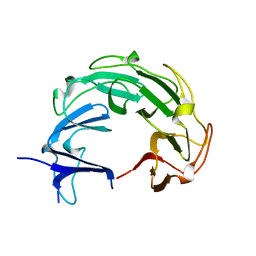 | |
2X4A
 
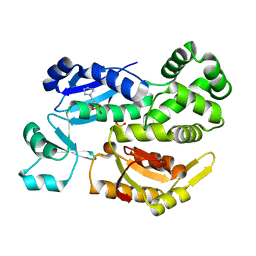 | |
5PZM
 
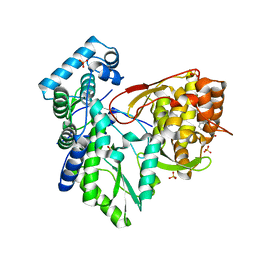 | |
2X49
 
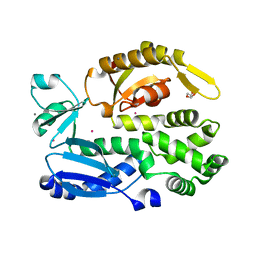 | | Crystal structure of the C-terminal domain of InvA | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, INVASION PROTEIN INVA, ... | | Authors: | Worrall, L.J, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the C-Terminal Domain of the Salmonella Type III Secretion System Export Apparatus Protein Inva.
Protein Sci., 19, 2010
|
|
3V46
 
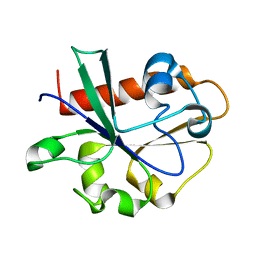 | | Crystal Structure of Yeast Cdc73 C-Terminal Domain | | Descriptor: | Cell division control protein 73 | | Authors: | Amrich, C.G, VanDemark, A.P. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Cdc73 subunit of Paf1 complex contains C-terminal Ras-like domain that promotes association of Paf1 complex with chromatin.
J.Biol.Chem., 287, 2012
|
|
1CSZ
 
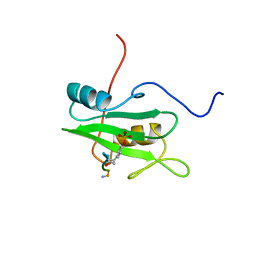 | | SYK TYROSINE KINASE C-TERMINAL SH2 DOMAIN COMPLEXED WITH A PHOSPHOPEPTIDEFROM THE GAMMA CHAIN OF THE HIGH AFFINITY IMMUNOGLOBIN G RECEPTOR, NMR | | Descriptor: | ACETYL-THR-PTR-GLU-THR-LEU-NH2, SYK PROTEIN TYROSINE KINASE | | Authors: | Narula, S.S, Yuan, R.W, Adams, S.E, Green, O.M, Green, J, Phillips, T.B, Zydowsky, L.D, Botfield, M.C, Hatada, M.H, Laird, E.R, Zoller, M.J, Karas, J.L, Dalgarno, D.C. | | Deposit date: | 1995-10-03 | | Release date: | 1996-11-08 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal SH2 domain of the human tyrosine kinase Syk complexed with a phosphotyrosine pentapeptide.
Structure, 3, 1995
|
|
2HL7
 
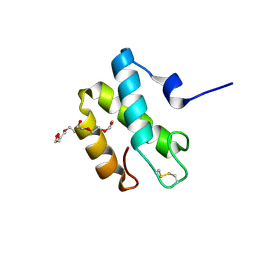 | | Crystal structure of the periplasmic domain of CcmH from Pseudomonas aeruginosa | | Descriptor: | Cytochrome C-type biogenesis protein CcmH, TETRAETHYLENE GLYCOL | | Authors: | Di Matteo, A, Travaglini-Allocatelli, C, Gianni, S, Brunori, M. | | Deposit date: | 2006-07-06 | | Release date: | 2007-07-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A strategic protein in cytochrome c maturation: three-dimensional structure of CcmH and binding to apocytochrome c
J.Biol.Chem., 282, 2007
|
|
5PZL
 
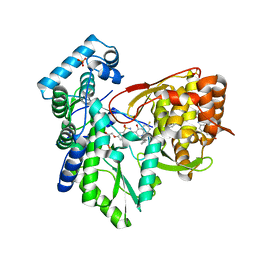 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE IN COMPLEX WITH 2-({3-[1-(2-CYCLOPROPYLETHYL)-6-FLUORO-4-HYDROXY-2-OXO-1,2-DIHYDROQUINOLIN-3-YL]-1,1-DIOXO-1,4-DIHYDRO-1LAMBDA~6~,2,4-BENZOTHIADIAZIN-7-YL}OXY)ACETAMIDE | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, 2-({3-[1-(2-cyclopropylethyl)-6-fluoro-4-hydroxy-2-oxo-1,2-dihydroquinolin-3-yl]-1,1-dioxo-1,4-dihydro-1lambda~6~,2,4-benzothiadiazin-7-yl}oxy)acetamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-27 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Discovery of a Hepatitis C Virus NS5B Replicase Palm Site Allosteric Inhibitor (BMS-929075) Advanced to Phase 1 Clinical Studies.
J. Med. Chem., 60, 2017
|
|
5PZK
 
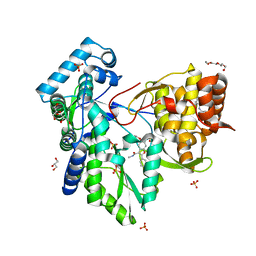 | |
4I33
 
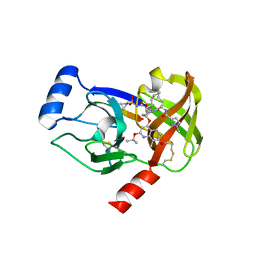 | | Crystal structure of HCV NS3/4A R155K protease complexed with compound 4 | | Descriptor: | (2R,6S,7E,10E,13aR,14aR,16aS)-2-{[7-methoxy-8-methyl-2-(propan-2-yloxy)quinolin-4-yl]oxy}-N-[(1-methylcyclopropyl)sulfonyl]-6-{[(1-methyl-1H-pyrazol-3-yl)carbonyl]amino}-5,16-dioxo-1,2,3,6,9,12,13,13a,14,15,16,16a-dodecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecine-14a(5H)-carboxamide, Genome polyprotein, HCV non-structural protein 4A, ... | | Authors: | Lemke, C.T. | | Deposit date: | 2012-11-23 | | Release date: | 2013-01-02 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.9001 Å) | | Cite: | Molecular Mechanism by Which a Potent Hepatitis C Virus NS3-NS4A Protease Inhibitor Overcomes Emergence of Resistance.
J.Biol.Chem., 288, 2013
|
|
3SU2
 
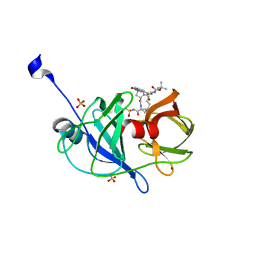 | | Crystal structure of NS3/4A protease variant A156T in complex with danoprevir | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, Genome polyprotein, SULFATE ION, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
5PZP
 
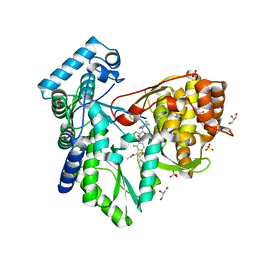 | |
3GS2
 
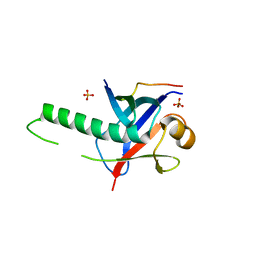 | | Ring1B C-terminal domain/Cbx7 Cbox Complex | | Descriptor: | Chromobox protein homolog 7, E3 ubiquitin-protein ligase RING2, SULFATE ION, ... | | Authors: | Wang, R, Taylor, A.B, Kim, C.A. | | Deposit date: | 2009-03-26 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Polycomb Group Targeting through Different Binding Partners of RING1B C-Terminal Domain.
Structure, 18, 2010
|
|
3RHK
 
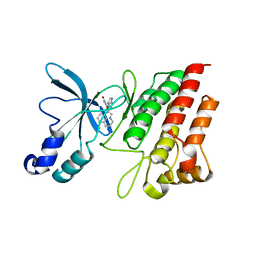 | | Crystal structure of the catalytic domain of c-Met kinase in complex with ARQ 197 | | Descriptor: | 1-[(3R,4R)-4-(1H-indol-3-yl)-2,5-dioxopyrrolidin-3-yl]pyrrolo[3,2,1-ij]quinolinium, Hepatocyte growth factor receptor | | Authors: | Eathiraj, S, Palma, R, Volckova, E, Hirschi, M, France, D.S, Ashwell, M.A, Chan, T.C. | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of a novel mode of protein kinase inhibition characterized by the mechanism of inhibition of human mesenchymal-epithelial transition factor (c-Met) protein autophosphorylation by ARQ 197.
J.Biol.Chem., 286, 2011
|
|
2PX7
 
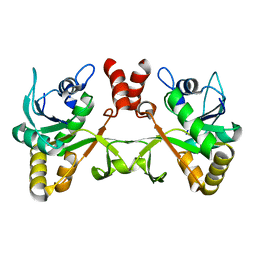 | | Crystal structure of 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase from Thermus thermophilus HB8 | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase | | Authors: | Chen, L, Tsukuda, M, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Chen, L.-Q, Liu, Z.-J, Lee, D, Chang, S.-H, Nguyen, D, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-14 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase from Thermus thermophilus HB8.
To be Published
|
|
